- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Localisation and Quantification of Reactive Oxygen Species and Nitric Oxide in Arabidopsis Roots in Response to Fungal Infection
Published: Vol 4, Iss 19, Oct 5, 2014 DOI: 10.21769/BioProtoc.1259 Views: 12536
Reviewed by: Igor Cesarino Zhaohui LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measurement of ATP Hydrolytic Activity of Plasma Membrane H+-ATPase from Arabidopsis thaliana Leaves
Masaki Okumura and Toshinori Kinoshita
Dec 5, 2016 9968 Views

Detection of Reactive Oxygen Species in Oryza sativa L. (Rice)
Navdeep Kaur [...] Pratap Kumar Pati
Dec 20, 2016 23856 Views

Evaluation of Root pH Change Through Gel Containing pH-sensitive Indicator Bromocresol Purple
Aparecida L. Silva [...] Daniel S. Moura
Apr 5, 2018 10022 Views
Abstract
Nitric oxide and reactive oxygen species have emerged as important signalling molecules in plants. The half-lives of NO and ROS are very short therefore rapid and precise measurements are required for the understanding biological roles of these redox active species. Various organelles and compartments generate NO and ROS thus it is important to determine precise location of these free radicals in order to understand their signalling roles. Diaminofluorescen (DAF) and fluorescent 2', 7'-dichlorofluorescein (DCF) dyes are employed to determine NO and ROS localisation. The advantage of this approach is that the dyes diffuse precisely to NO and ROS producing sites and generate fluorescence which can be detected by fluorescence- or confocal laser scanning microscopes. However, this technique has its disadvantages; particularly the specificity of the fluorescence signals needs to be established. Therefore, the use scavenger of NO such as cPTIO and ROS such as ascorbate is required to confirm the specificity of the fluorescence signal and ideally, confirmation of data obtained using other methods due to advantage and disadvantage associated with each method (Gupta and Igamberdiev, 2013). Here we describe a method to detect NO and ROS production from Arabidopsis roots in response to infection by Trichoderma, Fusarium using DAF, gas phase Griess reagent assay and DCF fluorescence methods.
Materials and Reagents
- Plant materials: 1-3 weeks old Arabidopsis thaliana sterile seedlings grown in vitro conditions
- 20 mM HEPES (pH 7.2) (Sigma-Aldrich, catalog number: H3375 )
- MS medium with vitamins (Duchefa Biochemie, catalog number: M0222 )
- Sodium hypochlorite (NaOCl) (Sigma-Aldrich, catalog number: 425044 )
- DAF-2DA (Enzo Life Sciences, catalog number: ALX-620-056-M001 )
- PDA medium (Potato dextrose agar)
- DCF-2DA fluorescent dye (Life Technologies, InvitrogenTM, catalog number: D-399 )
- Carboxy-PTIO potassium salt (Sigma-Aldrich, catalog number: C221 )
- Sulphaniliamide (Sigma-Aldrich, catalog number: S9251 )
- N-(1-naphthyl) ethylenediamine (NED) (Sigma-Aldrich, catalog number: 222488 )
- Potato dextrose agar (PDA) (Difco)
Equipment
- Slides
- Coverslips
- Leica fluorescent microscope
- Sterilised forceps
- Sterile pipette tips
- Micro centrifuge tubes
- Micropore tape (VWR International, catalog number: 115-8172 )
- Petri dishes
Software
- Image J software (version 1.45) Wayne Rasband (NIH)
Procedure
- Localisation of NO and ROS by DAF and DCF fluorescence
- Surface sterilize seeds of Arabidopsis thaliana (L.) Heynh. (Col-0) by incubating seeds in 10% NaOCl and 0.1% Tween-20 detergent for 10 min and washed three times with autoclaved distilled water.
- Transfer the sterilised seeds horizontally on Murashige Skoog (MS medium) 1.5% agar plates, close the plates with micropore tape and culture for 12 days under long day conditions .
- Grow Trichoderma asperelloides (T. asperelloides) T203 and Fusarium oxysporum (F. oxysporum) f. sp. lentis on potato dextrose agar (PDA; 39 g/L) plates for 10 days at 28 °C.
- Harvest T203 conidia by gentle scraping the petridish in 5 ml sterile water, consisting of both mycelium and spores, and add the solution (that contain mycelium+spores) to a 2-ml microcentrifuge tube in which the plant intact root system has to be placed (shown in Figure 1).
- The following treatments are recommended:
- Untreated plants (control).
- Incubate with T. asperelloides for various time intervals as described by Gupta et al. (2014).
- Incubate F. oxysporum-for 10 min to 4 h depending on experiment.
- Incubate plants simultaneously with T. asperelloides sand F. oxysporum.
- Untreated plants (control).
- Once the fungal incubation is finished remove plant from microcentrifuge and wash three times with HEPES buffer.
- Incubate the whole plant with 10 µM DAF-2DA or DCF-2DA (2 ml volume) for 10 min in dark (Figure 1).
- Wash excess of DAF or DCF-2DA dye three times with HEPES buffer.
- Observe roots under bright field mode and capture picture.
- The observe fluorescence emission using a 505- to 530-nm band-excitation filter coupled with a 515-nm long-emission filter.
Quantify images using Image J software by selecting area of interest and go to menu under Image J and click for analyse and then click submenu to measure.
- Surface sterilize seeds of Arabidopsis thaliana (L.) Heynh. (Col-0) by incubating seeds in 10% NaOCl and 0.1% Tween-20 detergent for 10 min and washed three times with autoclaved distilled water.
- Determination of NO by gas phase griess reagent assay
- Place 1 g of root material in a 10 ml small conical flask that contain 4 ml of HEPES buffer (Figure 1).
- Take a loop-full of inoculum, that contains both mycelium and spores, and add to a conical flask.
- The following treatments are recommended.
- Untreated plants (control).
- Incubate with T. asperelloides for various intervals.
- Incubate F. oxysporum-for various Intervals Figure 1.
- Incubate plants simultaneously with T. asperelloides and F. oxysporum.
- Untreated plants (control).
- Flush the solution in conical flask with NO free air (It is a safe process) (shown in Figure 1B).
- Trap the emitted air (bubbling) containing NO into 2 ml solution containing 1 ml of 1% w/v sulphanilamide and 0.02% w/v N-(1)-(naphthyl) ethylene-diaminedihydrochloride (Greiss reagent) (shown in Figure 1B).
- Read the absorbance at 540 nm in spectrophotometer.
- Make a standard curve with 0.1 to 5 µM nitrite.
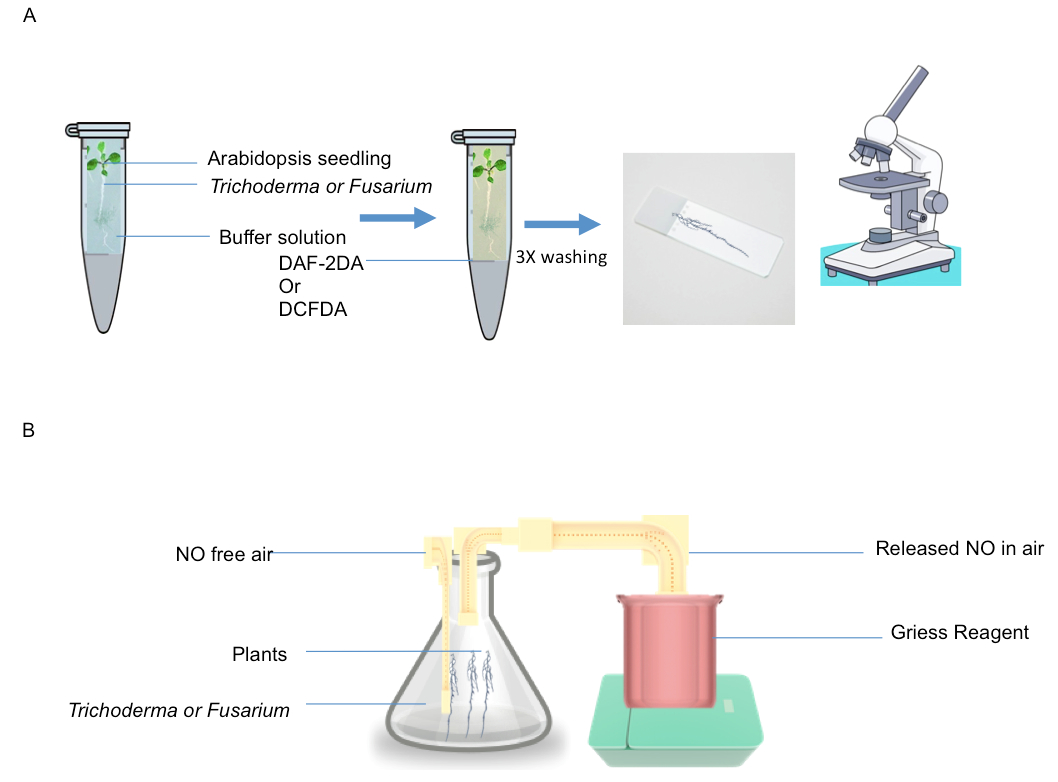
Figure1. A. NO and ROS detection by DAF and DCF2DA fluorescence; Roots were incubated in the solution containing fungus+spores and then transferred to DAF or DCF solution and monitored fluorescence B. Detection of NO released into gas phase detected by Griess reagent assay. 1 g of roots were placed in conical flask containing 4 ml of HEPES (pH 7.6) and flushed with NO free air and then NO generated from roots in response to fungal infection was trapped in griess reagent.
- Place 1 g of root material in a 10 ml small conical flask that contain 4 ml of HEPES buffer (Figure 1).
Acknowledgments
This work was supported by Max Planck Society (K. J. Gupta; Y. Brotman) and Marie Curie Intra European Fellowship for Carrier Development within the 7th European Community Framework Programme (K. J. Gupta) and the BBSRC-DEFRA-HGCA SCORE LINK grant (L. A. J. Mur).
References
- Gupta, K. J., Mur, L. A. and Brotman, Y. (2014). Trichoderma asperelloides suppresses nitric oxide generation elicited by Fusarium oxysporum in Arabidopsis roots. Mol Plant Microbe Interact 27(4): 307-314.
- Gupta, K. J. and Igamberdiev, A. U. (2013). Recommendations of using at least two different methods for measuring NO. Fron Plant Sci 4.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Gupta, K. J., Brotman, Y. and Mur, L. A. J. (2014). Localisation and Quantification of Reactive Oxygen Species and Nitric Oxide in Arabidopsis Roots in Response to Fungal Infection. Bio-protocol 4(19): e1259. DOI: 10.21769/BioProtoc.1259.
Category
Plant Science > Plant immunity > Perception and signaling
Plant Science > Plant physiology > Ion analysis
Biochemistry > Other compound > Reactive oxygen species
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









