- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detection of Hog1 Phosphorylation in Candida albicans in Response to an Antifungal Protein
Published: Vol 4, Iss 18, Sep 20, 2014 DOI: 10.21769/BioProtoc.1244 Views: 9338
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Far-western Blotting Detection of the Binding of Insulin Receptor Substrate to the Insulin Receptor
Jinghua Peng [...] Ling He
Feb 20, 2023 2493 Views

Use of a High-Affinity Ubiquitin-Binding Domain to Detect and Purify Ubiquitinated Substrates and Their Interacting Proteins
Nitu Saha [...] Mark Hochstrasser
Sep 5, 2025 3794 Views

Monitoring Endocytosis of Integral Membrane Proteins Using Western Blot-Based Detection of Biotinylated Antibody Uptake
Alexandra Graninger and Prasanna Satpute-Krishnan
Nov 20, 2025 2194 Views
Abstract
It is becoming increasingly apparent that stress signalling is important for tolerance of fungal species to antifungal chemicals and proteins. The high-osmolarity glycerol (HOG) pathway responds to a number of stressors including osmotic and oxidative stress. This protocol describes a method to detect activation of the Candida albicans (C. albicans) MAPK Hog1 by monitoring its phosphorylation in response to an antifungal protein.
Materials and Reagents
- Candida albicans wild-type (WT)
- C. albicans hog1Δ/Δ deletion mutant
- Half-strength potato dextrose broth (½ PDB) (BD, DifcoTM, catalog number: 254920 )
- Protein of interest (e.g. the plant defensin NaD1)
- 5 M NaCl
- Ultrapure ice-cold water
- Dry ice
- 20% trichloroacetic acid (TCA) (20% w/v)
- 10% trichloroacetic acid (TCA) (10% w/v)
- 100% ice-cold acetone
- 4x NuPAGE LDS sample buffer (Life Technologies, catalog number: NP0007 )
- Bond-Breaker TCEP solution (neutral pH) (Thermo Fisher Scientific, catalog number: 77720 )
- 1 M Tris (pH 8.0)
- Any kD Mini-PROTEAN TGX precast polyacrylamide gels (15 well) (Bio-Rad Laboratories, catalog number: 456-9036 )
- 10x Tris/glycine/SDS running buffer (Bio-Rad Laboratories, catalog number: 161-0072 )
- 5% (w/v) skim milk in TBS
- Phospho-p38 MAPK (Thr180/Tyr182) antibody (Cell Signaling Technology, catalog number: 9211 )
- Hog1 (y-215) antibody (Santa Cruz, catalog number: sc-9079 )
- Amersham ECL rabbit IgG (HRP-linked antibody from donkey) (GE, catalog number: NA934 )
- Amersham ECL Western blotting detection reagents (GE, catalog number: RPN2106 )
- Yeast peptone dextrose broth (YPD) (see Recipes)
- Yeast peptone dextrose agar (YPD agar) (see Recipes)
- Tris-buffered saline (TBS) (see Recipes)
- Tris-buffered saline with Tween 20 (TBST) (see Recipes)
Equipment
- 180 μm glass beads (acid-washed) (Sigma-Aldrich, catalog number: G1152 )
- Trans-blot turbo mini PVDF transfer pack (Bio-Rad Laboratories, catalog number: 170-4156 )
- 50 ml tubes screw-cap tubes
- 30 °C shaking incubator
- Spectrophotometer
- 500 ml baffled flask
- Eppendorf 5810 R centrifuge (Eppendorf, catalog number: 5811 000.010 )
- 1.5 ml microfuge tubes
- Vortex
- Bench top centrifuge
- Trans-blot turbo transfer system (Bio-Rad Laboratories, catalog number: 170-4155 )
- Circular mixer
- Bio-Rad ChemiDoc MP imaging system (Bio-Rad Laboratories, catalog number: 170-8280 )
Procedure
- Treatment of cells
- Plate C. albicans wild-type and hog1Δ strains onto YPD agar and grow overnight at 30 °C.
- Take scraping of both C. albicans strains from agar and inoculate 5 ml YPD in a 50 ml tube. Incubate tube overnight at 30 °C (250 rpm).
- Dilute both cultures to an OD600 of 0.2 (in 100 ml ½ PDB) and transfer 100 ml to a 500 ml baffled flask.
- Incubate cells at 30 °C (250 rpm) for 5 h (or until they reach an OD600 of 1.0).
- Dilute cells to an OD600 of 1.0. Transfer 10 ml aliquots of each culture to 50 ml tubes. One tube will be required per treatment (see Table 1 for treatment conditions).
- Treat 10 ml aliquots of WT and hog1Δ with NaCl or protein of interest (e.g. 10 μM and 20 μM NaD1 in ultrapure water). Cells will be incubated for 5, 10 or 15 min at 30 °C (250 rpm). For NaCl treatment, add 2.5 ml of 5 M NaCl to the 10 ml of WT or hog1Δ.
For protein treatments, add volume to reach desired protein concentration (e.g. volume of protein for a final concentration of 10 μM or 20 μM). For the hog1∆ control treatments, only the 20 µM treatment concentration is required. If no total Hog1 or phosphorylated Hog1 is detected in the hog1∆ cells treated with 20 µM protein, then none will be detectable at a lower protein concentration.
Table 1. Treatment conditions. Treatment conditions of C. albicans WT and hog1∆ cells with the protein of interest. 1 M NaCl is included as a positive control. No-protein and hog1∆ treatments are included as negative controls.C. albicans strain Protein or NaCl concentration Incubation time WT No treatment - WT 1 M NaCl 10 min WT 10 μM protein 5 min WT 10 μM protein 10 min WT 10 μM protein 15 min WT 20 μM protein 5 min WT 20 μM protein 10 min WT 20 μM protein 15 min hog1Δ No treatment - hog1Δ 1 M NaCl 10 min hog1Δ 20 μM protein 5 min hog1Δ 20 μM protein 10 min hog1Δ 20 μM protein 15 min - After incubation spin cells down at 1,800 x g for 5 min (at RT). Discard the supernatant.
- Resuspend cells in 500 μl ice-cold water. Transfer to a 1.5 ml microfuge tube (Tube A).
- Centrifuge cells at 3,000 x g for 5 min (at RT). Discard the supernatant.
- Freeze pellets with dry ice.
- Plate C. albicans wild-type and hog1Δ strains onto YPD agar and grow overnight at 30 °C.
- Protein preparation
- Resuspend cell pellets in 100 μl 20% TCA. Also add approximately 100 μl of 180 μm glass beads. Keep pellets on dry ice.
- Vortex tubes for 2 min (at highest speed).
- Transfer supernatant from Tube A to a new tube (Tube B).
- Wash the remaining beads in Tube A with 500 μl 10% TCA. Transfer the 500 μl TCA and add it to Tube B.
- Spin Tube B at 18,000 x g for 5 min (at RT). Remove supernatant.
- Resuspend pellets in 1 ml ice-cold acetone.
- Spin again at 18,000 x g for 5 min (at RT). Remove supernatant.
- Resuspend pellets with 50 μl loading buffer (1x NuPAGE LDS sample buffer and 50 mM Bond-Breaker). If the buffer turns yellow, add a few microliters of 1 M Tris (pH 8.0) until the color becomes blue.
- Heat samples at 70 °C for 10 min.
- Resuspend cell pellets in 100 μl 20% TCA. Also add approximately 100 μl of 180 μm glass beads. Keep pellets on dry ice.
- SDS-PAGE and Western blots
- Load 25 μl of each sample into the wells of a Bio-Rad any kD Mini-PROTEAN TGX precast polyacrylamide gels, 15 well (Gel 1). Repeat this for a second gel (Gel 2).
- Run gel for 30 min at 200 V (with 1x Tris/Glycine/SDS running buffer).
- Transfer protein to Trans-blot turbo mini PVDF membranes (from trans-blot turbo mini PVDF transfer pack) using the Bio-Rad trans-blot turbo transfer system (1.3 A, 25 V, 7 min).
- Block PVDF membranes in 5% skim milk (40 ml TBS, in a 50 ml tube) for 2 h at RT (on a circular mixer). Discard skim milk after incubation.
- Add primary antibodies and incubate overnight on a circular mixer at 4 °C (in a 50 ml tube).
For Gel 1, dilute Hog1 (y-215) antibody to 1 in 200 with 10 ml 5% skim milk in TBST.
For Gel 2, dilute Phospho-p38 MAPK (Thr180/Tyr182) antibody to 1 in 200 with 10 ml 5% skim milk in TBST. - Remove primary antibodies. Wash membranes with TBST (3x, 10 min per wash).
- Dilute secondary antibody (ECL rabbit IgG, HRP-linked) to 1 in 20,000 with 10 ml TBST and incubate with membranes overnight at 4 °C.
- Remove secondary antibodies. Wash membranes with TBST (3x, 10 min per wash).
- Mix 1 ml ECL detection reagent 1 and 1 ml of ECL detection reagent 2 for each membrane. Add 2 ml of the mixture to each membrane.
- Expose membranes using chemiluminescent settings on a Bio-Rad ChemiDoc MP imaging system. Expose membranes for ~ 1 min (or longer if the signal is weak).
See Figure 1 for an example.
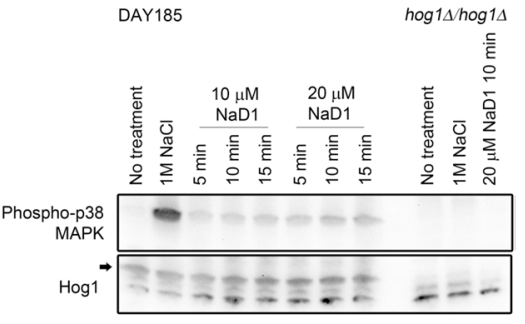
Figure 1. Treatment of C. albicans with NaD1 leads to Hog1 phosphorylation. Western blots showing C. albicans Hog1 phosphorylation (detected using Phospho-p38 MAPK antibody) after treatment with the plant defensin NaD1. Bands corresponding to total Hog1 levels are indicated with an arrow. Total Hog1 levels are consistent across all WT treatments and are absent from the hog1∆ mutant. No phosphorylated Hog1 is detected in the no treatment or hog1∆ negative controls. Hog1 phosphorylation is detected in the 1 M NaCl positive control and in all NaD1 treated samples. In this example only one time point is shown for the 20 µM NaD1 treated hog1∆ control. Image originally published in Reference 1 (Hayes et al., 2013). - Load 25 μl of each sample into the wells of a Bio-Rad any kD Mini-PROTEAN TGX precast polyacrylamide gels, 15 well (Gel 1). Repeat this for a second gel (Gel 2).
Recipes
- YPD or YPD agar
1% yeast extract
2% peptone
2% dextrose
If making plates also add 2% agar - TBS and TBST
400 mM Tris (pH 7.5)
3 M Tris
For TBST also add 0.1% Tween-20
Acknowledgments
This protocol has been adapted from Hayes et al. (2013), in which a brief description of the protocol was presented.
References
- Hayes, B. M., Bleackley, M. R., Wiltshire, J. L., Anderson, M. A., Traven, A. and van der Weerden, N. L. (2013). Identification and mechanism of action of the plant defensin NaD1 as a new member of the antifungal drug arsenal against Candida albicans. Antimicrob Agents Chemother 57(8): 3667-3675.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hayes, B. M. and Weerden, N. L. V. D. (2014). Detection of Hog1 Phosphorylation in Candida albicans in Response to an Antifungal Protein. Bio-protocol 4(18): e1244. DOI: 10.21769/BioProtoc.1244.
Category
Microbiology > Antimicrobial assay > Antifungal assay
Microbiology > Microbial signaling > Phosphorylation
Biochemistry > Protein > Immunodetection > Western blot
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









