- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protocol for Biotin Bioassay-based Cross Feeding
Published: Vol 4, Iss 18, Sep 20, 2014 DOI: 10.21769/BioProtoc.1242 Views: 8113
Reviewed by: Kanika GeraAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

β-lactamase (Bla) Reporter-based System to Study Flagellar Type 3 Secretion in Salmonella
Fabienne F. V. Chevance and Kelly T. Hughes
Jun 20, 2023 1775 Views

Determination of Poly(3-hydroxybutyrate) Content in Cyanobacterium Synechocystis sp. PCC 6803 Using Acid Hydrolysis Followed by High-performance Liquid Chromatography
Janine Kaewbai-ngam [...] Tanakarn Monshupanee
Aug 20, 2023 1825 Views

An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis
Avisek Mahapa [...] Dipankar Chatterji
Dec 20, 2024 1790 Views
Abstract
Biotin bioassay-based cross-feeding experiments were performed to elucidate the effect on biotin production by bioRbme expression in Agrobacterium tumefaciens (A. tumefaciens) (Feng et al., 2013). The indicator strain used here is the biotin auxotrophic strain of Escherichia coli (E. coli), ER90 (ΔbioF bioC bioD), which was cross-fed by A. tumefaciens species (Feng et al., 2013a). The biotin-free M9 minimal medium plates were formulated as described by other and our research groups (Feng et al., 2013b; Lin et al., 2010; del et al., 1979). Of note, 0.01% (w/v) the redox indicator 2, 3, 5-triphenyl tetrazolium chloride (TTC) was supplemented into the above media. Consequently, biotin generation/production was observed via the reduction of TTC to the insoluble red formazan which is due to the ER90 growth fed by A. tumefaciens strains (Feng et al., 2014). Detailed procedures are described as follows.
Materials and Reagents
- Four A. tumefaciens strains [NTL4 (WT), FYJ283 (ΔbioBFDA), FYJ212 (ΔbioRat) and FYJ341 (ΔbioR::Km+bioRbme)]
- Biotin auxotrophic strain of E. coli, ER90
- 1 nM biotin (Sigma-Aldrich, catalog number: B4510 )
- 0.01% (w/v) 2, 3, 5-triphenyl tetrazolium chloride (TTC) (AMRESCO, catalog number: 0 765 )
- 0.1% vitamin-free casamino acids hydrolysate (Sigma-Aldrich, catalog number: C7710 )
- MgSO4
- Glucose
- M9 minimal medium (see Recipes)
Equipment
- Centrifuge
- Petri dishes Petri dishes (90 mm) (Thermo Fisher Scientific, catalog number: 502VF )
- Sterile paper disks (6 mm, BBL)
Procedure
- Preparation of biotin bioassay plates
- The biotin assay plates were prepared as previously described with few changes, one of which referred to the thinner thickness of the plate agar where we dropped paper discs (Feng et al., 2013b; Lin et al., 2010).
- Overnight cultures of strain ER90 (grown in 6 ml of defined M9 minimal medium with 1 nM biotin at 30 °C) were collected by centrifugation (3,600 rpm, 16 min), washed three times with the same volume (6 ml) of M9 medium, and were cultivated at 37 °C to 0.8 OD600 in 200 ml of M9 minimal medium containing 1 nM biotin.
- To remove excess of biotin, all the bacterial cells from 200 ml culture were washed twice in M9 media and sub-cultured into 1 L of M9 minimal medium at 37 °C for 5 h to de-repress expression of bio operon by starvation for biotin.
- The bacteria were harvested by centrifugation (3,600 rpm, 16 min), washed three times with M9 medium, re-suspended in 1 ml of the same medium and mixed into 150 ml of the defined M9 agar media supplemented with 0.01% (w/v) TTC as a redox indicator.
- Finally, the mixture (5 ml per sector) was poured into Petri dishes sectored with plastic walls to avoid cross-feeding and a sterile paper disk (6 mm, BBL) was centered on the agar top of each sector.
- The biotin assay plates were prepared as previously described with few changes, one of which referred to the thinner thickness of the plate agar where we dropped paper discs (Feng et al., 2013b; Lin et al., 2010).
- Preparation of cross-feeder strains
- In total, four feeder strains of A. tumefaciens corresponded to NTL4 (WT), FYJ283. (ΔbioBFDA), FYJ212 (ΔbioRat) and FYJ341 (ΔbioR::Km+bioRbme).
- The biotin auxotroph strain FYJ283 was cultivated in 5 ml of M9 medium supplemented with 1 nM biotin, whereas the other three strains were cultivated in 5 ml of biotin-free M9 minimal media overnight.
- Overnight cultures were collected by centrifugation (3,000 rpm, 10 min), washed three times using the M9 liquid medium, and transferred into 100 ml of biotin-free M9 media for 6 more hours of growth at 30 °C to deplete trace amounts of intracellular biotin in the biotin auxotroph strain FYJ283.
- Following three rounds of washing with same media, bacteria were resuspended in M9 media and their optical densities at 600 nM (OD600) were adjusted to 1.5. 20 μl of A. tumefaciens culture (OD600 = 1.0) was spotted on the paper disc, and incubated overnight at 30 °C.
- Finally, the red deposit of formazan (Lin et al., 2010; del et al., 1979; Feng et al., 2014) suggests that the indicator strain ER90 is fed by the A. tumefaciens strains (seen in Figure 1), and the area size (square centimeters) of formazan represents the level of biotin pool produced by the different feeder strains.
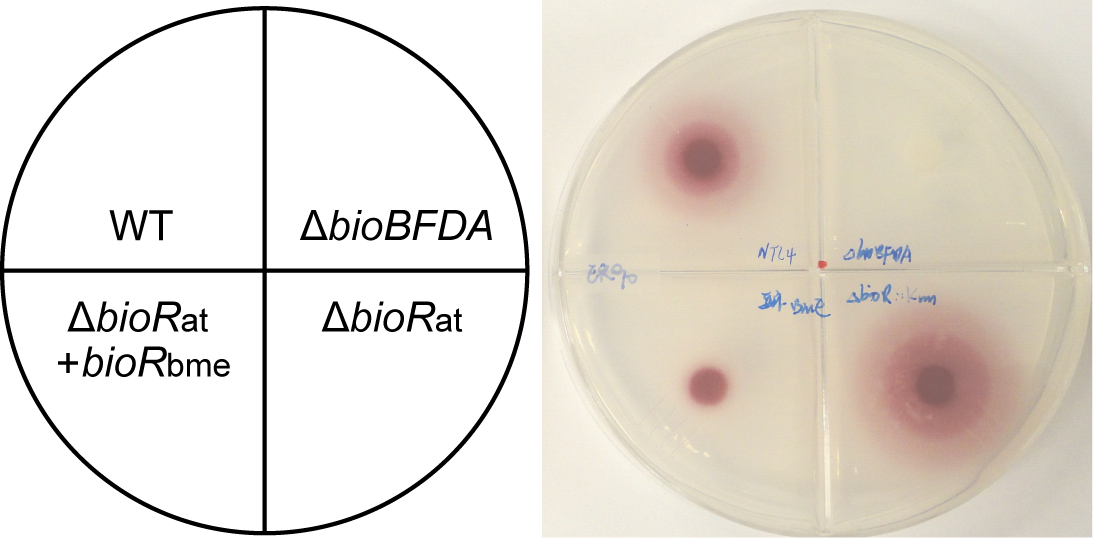
Figure 1. A representative photograph illustrating the relevance of BioR-mediated regulation to biotin synthesis. It was fully adapted from Feng et al. (2013a). Four A. tumefaciens strains cross-feed E. coli strain ER90 with deletion of full bio operon, which are NTL4 (WT), FYJ283 (ΔbioBFDA), FYJ212 (ΔbioRat), and FYJ 341 (ΔbioRat+bioRbme), respectively.
- In total, four feeder strains of A. tumefaciens corresponded to NTL4 (WT), FYJ283. (ΔbioBFDA), FYJ212 (ΔbioRat) and FYJ341 (ΔbioR::Km+bioRbme).
Recipes
- M9 minimal medium
6 g of Na2PO4, 3 g of KH2PO4, 0.5 g of NaCl and 1 g of NH4Cl per liter
Acknowledgments
This protocol was adapted/modified from previous works seen in del Campillo-Campbell et al. (1079); Feng et al. (2014); Feng et al. (2013a); Feng et al. (2013b) and Lin et al. (2010).
References
- del Campillo-Campbell, A., Dykhuizen, D. and Cleary, P. P. (1979). Enzymic reduction of d-biotin d-sulfoxide to d-biotin. Methods Enzymol 62: 379-385.
- Feng, Y., Napier, B. A., Manandhar, M., Henke, S. K., Weiss, D. S. and Cronan, J. E. (2014). A Francisella virulence factor catalyses an essential reaction of biotin synthesis. Mol Microbiol 91(2): 300-314.
- Feng, Y., Xu, J., Zhang, H., Chen, Z. and Srinivas, S. (2013a). Brucella BioR regulator defines a complex regulatory mechanism for bacterial biotin metabolism. J Bacteriol 195(15): 3451-3467.
- Feng, Y., Zhang, H. and Cronan, J. E. (2013b). Profligate biotin synthesis in α‐proteobacteria–a developing or degenerating regulatory system? Mol Microbiol 88(1): 77-92.
- Lin, S., Hanson, R. E. and Cronan, J. E. (2010). Biotin synthesis begins by hijacking the fatty acid synthetic pathway. Nat Chem Biol 6(9): 682-688.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Feng, Y., Xu, J., Zhang, H., Chen, Z. and Srinivas, S. (2014). Protocol for Biotin Bioassay-based Cross Feeding. Bio-protocol 4(18): e1242. DOI: 10.21769/BioProtoc.1242.
Category
Microbiology > Microbial metabolism > Other compound
Microbiology > Microbial biochemistry > Other compound
Microbiology > Microbial cell biology > Cell staining
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









