- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Activity Assays for Bacteriophage Endolysin PlyPy
Published: Vol 4, Iss 18, Sep 20, 2014 DOI: 10.21769/BioProtoc.1233 Views: 11078
Reviewed by: Kanika GeraAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

H2 Production from Methyl Viologen–Dependent Hydrogenase Activity Monitored by Gas Chromatography
Nuttavut Kosem
Dec 5, 2023 1770 Views

Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
John S. Smetana [...] Christopher W. Lennon
Apr 20, 2025 1585 Views

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1928 Views
Abstract
Bacterial viruses (bacteriophages) escape and kill their host by degrading the bacterial peptidoglycan layer through the mechanism of enzymes called endolysins: peptidoglycan degrading enzymes. The method included here is useful for the initial characterization of any endolysin, regardless of the specific catalytic domain (as long as the activity results in a reduction in the optical density), in order to determine its optimal enzymatic (lytic) activity. This protocol is specific for the Streptococcus pyogenes phage endolysin PlyPy, but can be adapted for any peptidoglycan degrading enzyme.
Keywords: Phage lysinsMaterials and Reagents
- Streptococcus pyogenes (S. pyogenes) strain MGAS 3 15
- Purified phage lysin PlyPy (or other lysins of interest)
- Brain heart infusion (BHI) broth (BD, catalog number: 237500 )
- Agar (BD, catalog number: 214010 )
- Tris (Thermo Fisher Scientific, catalog number: BP152-1 )
- HCl (Thermo Fisher Scientific, catalog number: A144S-500 )
- CaCl2.2H2O (Thermo Fisher Scientific, catalog number: C70-500 )
- MgCl2.7H2O (AMRESCO, catalog number: 0288-100G )
- ZnCl2 (Fluka, catalog number: 96468-50G )
- DTT (Sigma-Aldrich, catalog number: D0632-10G )
- Sodium acetate (Thermo Fisher Scientific, catalog number: S210-500 )
- NaCl (Thermo Fisher Scientific, catalog number: S271-3 )
- PlyPy activity buffer (see Recipes)
Equipment
- 37 °C incubator
- Spectrophotometer (Bio-Rad Laboratories, SmartSpecTM, model: 3000 )
- Table centrifuge (Eppendorf, model: 5810 R )
- pH meter (Thermo Fisher Scientific, AccumetTM, model: 15)
- 15 ml tubes (Falcon®, catalog number: 352097 )
- 50 ml tubes (Falcon®, catalog number: 352098 )
- Cuvettes (Sarstedt AG, catalog number: 67.742 )
- 96-well flat bottom plates (Falcon®, catalog number: 35-1172 )
- 96-well plate reader (Molecular Devices, SpectraMax Plus Reader)
Procedure
Note: Streak S. pyogenes strain MGAS 315 on a BHI agar plate (or other plates supporting the growth of your bacteria, including blood agar and THY agar plates) and allow to incubate overnight at 37 °C until single colonies are visible.
- Inoculate one colony of MGAS 315 in 5 ml BHI in a 15 ml tube. Incubate overnight at 37 °C.
- Preheat BHI by adding 45 ml BHI to a 50 ml tube, and incubate overnight at 37 °C.
- Transfer the 5 ml overnight culture of MGAS 315 into the preheated BHI (1:10 dilution), and incubate until OD600 ∼ 0.3. The time for the culture to reach this OD600 varies depending on what strain is used, but will in general take 3-4 h for S. pyogenes.
Note: Cells can be taken at other OD600 values, but bacteria are generally more susceptible to the effect of endogenously added lysins when in mid log phase as compared to stationary phase cells. Therefore it is recommended to do a growth curve of your bacteria to determine the mid log phase before conducting any further experiments. - Harvest cells by centrifugation (4,000 x g; 15 min; 4 °C).
- Discard supernatant, and wash cells three times in 20 mM Tris-HCl (pH 6.8) (4,000 x g; 15 min; 4 °C).
Note: Other buffers might prove more efficient for other peptidoglycan degrading enzymes, but buffers in the pH-range 6-8 are usually a good starting point since bacteria generally tolerate this pH well, and most lysins are highly active within this pH range. - Resuspend the washed cells in 20 mM Tris-HCl (pH 6.8) to an OD600 ∼ 1.4.
Note: For best experimental resolution, a final OD600 of 0.5-1.0 is recommended. Washed cells will be diluted 1:1 in upcoming steps, yielding a final OD600 = 0.7. - Add 100 μl washed cells to the wells of a sterile 96-well plate.
Note: For consistency and statistical reliability, we recommend all samples to be run in at least triplicates. - Add 100 μl buffer [20 mM Tris-HCl (pH 6.8)] containing divalent cations at varying concentrations.
CaCl2: 0-100 mM
MgCl2: 0-100 mM
ZnCl2: 0-5 mM
Note: Since many endolysins rely on the presence of divalent cations for their activity, it is critical to optimize this condition before moving on to other optimizations. Several other divalent cations might affect the activity of lysins, including but not limited to Mn2+, Fe2+, Co2+, Cu2+ and Ni2+. - Add 1 μg of your peptidoglycan hydrolyzing protein (PlyPy).
Note: The amount of lysin needed to be added is dependent on the activity of the lysin and needs to be determined empirically. A few μg lysin is a good starting point. - Insert the 96-well plate into the SpectraMax Plus Reader, and incubate the plate at 37 °C for 60 min. Shake the plate and measure the OD600 every 30 sec.
- Plot the data in an Excel sheet and analyze the results.
- Repeat steps 9-12, including the necessary divalent cations at an empirically determined concentration in the buffer, but now optimize the conditions for other factors, including but not limited to:
NaCl: 0-1,000 mM
DTT: 0-20 mM
Temperature: 20-50 °C - To determine the optimal pH, exchange the 20 mM Tris-HCl (pH 6.8) used in steps 6, 7 and 9 to buffers of your choice, including but not limited to:
20 mM sodium acetate (pH 5.0)
20 mM sodium acetate (pH 5.5)
20 mM sodium acetate (pH 6.0)
20 mM Tris-HCl (pH 6.0)
20 mM Tris-HCl (pH 6.5)
20 mM Tris-HCl (pH 7.0)
20 mM Tris-HCl (pH 7.5)
20 mM Tris-HCl (pH 8.0)
20 mM Tris-HCl (pH 8.8) - To finally determine the activity of your lysin:
- Wash and resuspend your cells in activity buffer (optimized buffer based on your empirical values) to an OD600 ∼ 1.4.
- Add 100 μl cells and 100 μl activity buffer to each well in a 96-well plate.
Note: Take into account that the maximum volume in the wells will be 200 μl. Therefore, less than 100 μl activity buffer will be added to wells that will also contain lysin (diluted in activity buffer). The exact volume depends on your initial concentration of the lysin. - Add your lysin in serial dilutions, ranging from 0.1-51.2 μg/ml.
Note: It is important to run all samples in triplicates to account for any outliers. - Incubate for 15 min at 37 °C in a SpectraPlus Max Reader, shaking and measuring OD600 of the samples every 30 sec.
- Plot the data in an Excel sheet and analyze the results.
- The dilution closest corresponding to a 50% reduction of the starting OD corresponds to 1 Unit (Loeffler et al., 2001). Most characterized lysins have an activity of 0.5-1 U/μg.
- Wash and resuspend your cells in activity buffer (optimized buffer based on your empirical values) to an OD600 ∼ 1.4.
Representative data
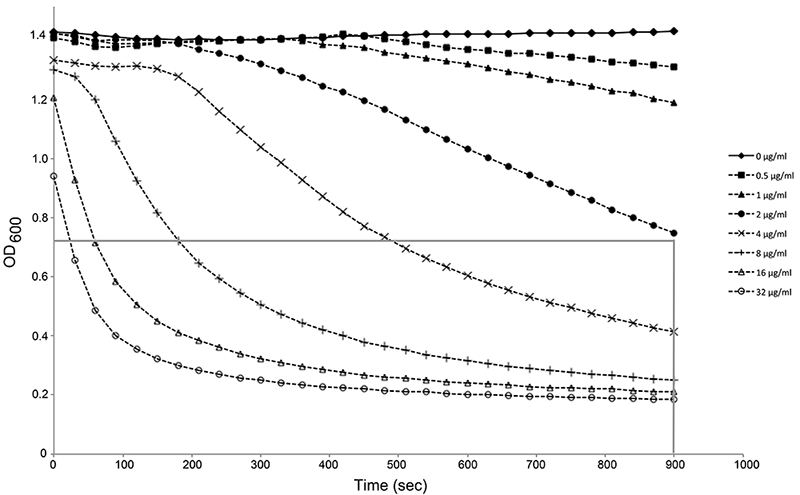
Figure 1. Activity determination of PlyPy. Washed S. pyogenes cells were incubated in activity buffer at 37 °C for 15 min with varying concentrations of PlyPy. The optical density (600) was measured every 30 sec in a SpectraPlus Max Reader. The mean value of triplicates is shown. For simplicity, a gray line has been added, representing the OD50% at 900 sec (15 min), thus corresponding to one Unit. In this representative figure, 2 μg/ml corresponds closely to one Unit; e.g. the activity of the lysin is 0.5 U/μg. Note the fast initial drop in optical density in samples with high concentration of lysin, stressing the necessity of reading your sample as soon as possible after adding the lysin.
Recipes
- PlyPy activity buffer
20 mM Tris-HCl (pH 6.8)
2 mM CaCl2
100 mM NaCl
Sterile filter the solution (0.22 μm) before usage
Acknowledgments
This protocol was published in a more abbreviated form in the full-length article by Lood et al. (2014). This study was supported in part by USPHS grant AI11822 and by a grant from Contrafect Inc. to V.A.F. and by a grant from the Tegger Foundation to R.L.
References
- Loeffler, J. M., Nelson, D. and Fischetti, V. A. (2001). Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 294(5549): 2170-2172.
- Lood, R., Raz, A., Molina, H., Euler, C. W. and Fischetti, V. A. (2014). A highly active and negatively charged streptococcus pyogenes lysin with a rare d-alanyl-l-alanine endopeptidase activity protects mice against Streptococcal bacteremia. Antimicrob Agents Chemother 58(6): 3073-3084.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lood, R. and Fischetti, V. A. (2014). Activity Assays for Bacteriophage Endolysin PlyPy. Bio-protocol 4(18): e1233. DOI: 10.21769/BioProtoc.1233.
Category
Microbiology > Microbe-host interactions > Virus
Microbiology > Microbial biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











