- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Dye-uptake Experiment through Connexin Hemichannels
Published: Vol 4, Iss 17, Sep 5, 2014 DOI: 10.21769/BioProtoc.1221 Views: 11986
Reviewed by: Vanesa Olivares-IllanaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantifying Intracellular Distributions of HaloTag-Labeled Proteins With SDS-PAGE and Epifluorescence Microscopy
Julia Shangguan and Ronald S. Rock
Jul 20, 2025 2533 Views

Fluorescence Lifetime-Based Separation of FAST-Labeled Cellular Compartment
Aidar R. Gilvanov [...] Yulia A. Bogdanova
Oct 5, 2025 1350 Views

Purification of the Active-State G Protein-Coupled Receptor ADGRL4 for Cryo-Electron Microscopy Using a Modular Tag System and a Tethered mini-Gq
David M. Favara and Christopher G. Tate
Mar 5, 2026 140 Views
Abstract
Connexins (Cxs) are integral membrane proteins of vertebrates that associate to form hexameric transmembrane channels, named hemichannels. Twenty-one Cx types have been described, which are named according to their molecular weight. Cxs are expressed in many cell types, e.g. epithelial cells, astrocytes and immune cells. Hemichannels allow the passage of molecules of up to 1-2 kDa along the concentration gradient. When surface-exposed, hemichannels mediate the exchange of molecules between the cytosol and the extracellular space. Hemichannels are closed by default, but several cues inducing their opening have been described, e.g. a drop in the extracellular Ca2+ concentration (Evans et al., 2006) or infection with enteric pathogens (Puhar et al., 2013; Tran Van Nhieu et al., 2003). Hemichannel opening can be measured by electrophysiology, by quantifying the release of a hemichannel-permeable molecule into the extracellular medium or by quantifying the uptake of a hemichannel-permeable, plasma membrane-impermeant molecule. As the extent of uptake of a molecule is proportional to its concentration, exposure time, temperature (these parameters are controlled) and, importantly, to the number of active hemichannels on the cell surface, uptake assays are routinely used to assess hemichannel opening. This protocol for the uptake of the fluorescent dye ethidium bromide was used with Hela cells that were stably transfected with Cx26 or Cx43 (Paemeleire et al., 2000). Nevertheless, it could likely be used with other Cx-expressing cell types.
Materials and Reagents
- Cell line of interest and assorted cell culture media and plasticware for propagation from supplier of choice
- DMEM low glucose (Life Technologies, Gibco®, catalog number: A1515401 )
- 100x MEM non-essential amino acids (Life Technologies, Gibco®, catalog number: 11140050 )
- 100x 10,000 U/ml penicillin-streptomycin (Life Technologies, Gibco®, catalog number: 15140122 )
- Fetal bovine serum (FBS) (heat-inactivated) (Life Technologies, Gibco®, catalog number: 10500056 )
- Hank’s buffered salt solution (HBSS) (no phenol red) (Life Technologies, Gibco®, catalog number: 14025092 )
- Hank’s buffered salt solution (HBSS) (no calcium, no magnesium, no phenol red) (Life Technologies, Gibco®, catalog number: 14175095 )
- 10x HBSS stock solutions instead of ready to use buffer if desired (Life Technologies, Gibco®, catalog numbers: 14065056 and 14185052 )
- 1 M HEPES (Life Technologies, Gibco®, catalog number: 15630080 )
- 1 M MgCl2
- 0.5 M Na-EGTA
- Ethidium bromide solution (here: eurobio, 0.7 mg/ml)
- Culture medium for propagation of HelaCx26 or HelaCx43 cells (see Recipes)
- HBSS solution (see Recipes)
- HBSS solution devoid of Ca2+ (see Recipes)
- HBSS stock with ethidium bromide (see Recipes)
- Acid washed coverslips (see Recipes)
Equipment
- Incubator for cell culture
- Cover slips and holder or glass bottom dishes for live cell imaging (e.g. MatTek)
- Water bath
- Pipettes
- Aspiration
- Clean bench
- Fluorescence microscope (ideally temperature-controlled) equipped with a camera
Software
- An image analysis software [e.g. ImageJ (http://imagej.nih.gov/ij/) or MetaMorph (Molecular Devices)]
Procedure
- Cells were grown at 37 °C and 10% CO2 in a humid environment (incubator) and passaged according to standard protocols.
- Cells were seeded on acid-washed cover slips of desired size (e.g. 24 well plates) (see Recipes) or glass-bottom dishes the day before the experiment and used at 70 % confluency.
- A stock solution of HBSS devoid of Ca2+ containing ethidium bromide (e.g. 5 μM) was prepared to make sure that all samples were exposed to exactly the same concentration of dye.
- Cells were carefully washed twice with warm (37 °C) HBSS and once with warm HBSS devoid of Ca2+. For negative controls, only HBSS was used. To wash, carefully aspirate the medium or buffer, replace with fresh buffer, carefully pipet up and down once or twice, and replace with fresh buffer.
- Cells were loaded with pre-warmed 5 μM ethidium bromide in HBSS devoid of Ca2+ (to induce hemichannel opening) for 10 min at 37 °C. For negative controls pre-warmed 5 μM ethidium bromide in HBSS containing Ca2+ was used.
- The supernatant was discarded and cells washed with warm HBSS containing Ca2+ to induce hemichannel closure and thereby trap the dye in the cytosol.
- Images were acquired within 30 min (the ethidium bromide signal was stable during this period) at 37 °C and 40x magnification. Make sure enough separate fields within a sample are acquired and prepare sufficient independent repeats for quantification. Please refer to Notes for the settings of the fluorescence microscope.
- Intracellular ethidium bromide-fluorescence of single cells was quantified using an image analysis software. The background fluorescence was subtracted for every field.
Representative data
- Representative result
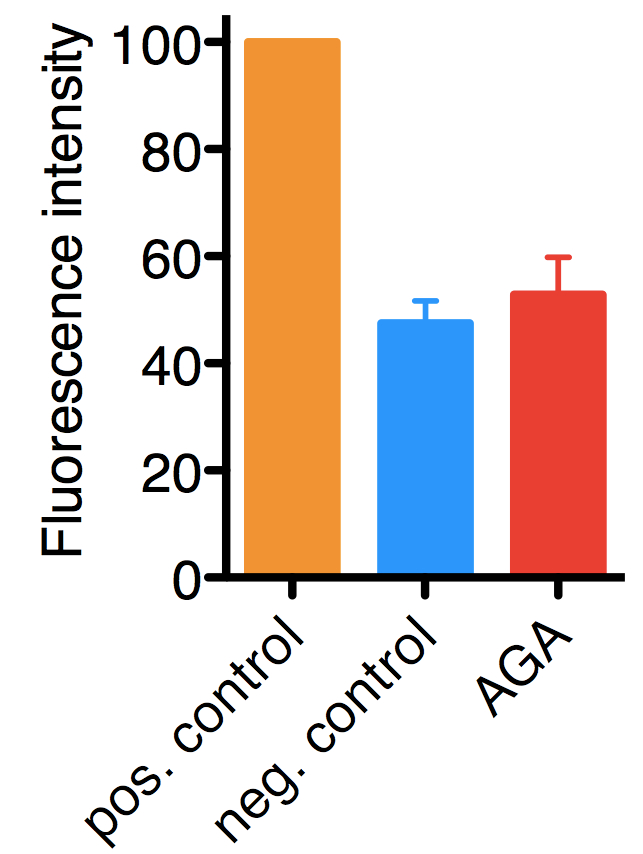
Figure 1. Typical results for the assay performed on HelaCx26 cells. Uptake of ethidium bromide was quantified by fluorescence and values were normalized to those of positive controls (in which hemichannel opening was induced by low extracellular Ca2+), which were set to 100. In negative controls hemichannel opening was not induced, hence, the background fluorescence corresponds mostly to ethidium bromide taken up by endocytosis. The mean with SD of approximately 900 cells from three independent experiments are shown.18α-glycyrrhetinic acid (AGA), 100 μM (a hemichannel inhibitor).
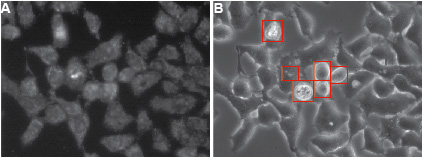
Figure 2. Typical appearance of HelaCx26 cells loaded with ethidium bromide. Cells were exposed for 10 min to ethidium bromide in HBSS devoid of Ca2+ to induce hemichannel opening (this type of sample corresponds to a positive control in Figure 1). Fluorescent A and transmission B images of cells are shown. Dying cells and debris (highlighted by red rectangles in B) should be omitted for quantification. The compromised plasma membrane permeability of dying cells will allow non-hemichannel-dependent passage of ethidium bromide. Ethidium bromide fluorescence is enhanced when the dye is bound to DNA.
- Notes about reproducibility and variability in results
Variability is low (10%), as dye uptake is very homogenous with a population of cells. There is an about two to three-fold difference between positive and negative controls. Shorter ethidium bromide pulses result in higher variability (due to differences in handling time), but also slightly higher differences between positive and negative controls, because less ethidium bromide is taken up by endocytosis.
Notes
- Please also refer to the protocol “Induction of Connexin-hemichannel Opening” described by Puhar and Sansonetti (2014).
- Many cell lines have lost expression of Cxs and therefore do not have hemichannels. Make sure your cell line of interest does.
- During the experiment, handle cells carefully, as hemichannels are mechanosensitive. Excessive shaking and vigorous exchange of media will induce hemichannel opening and therefore increase the background for subsequent measurements.
- For HBSS, 10x stock solutions may be used.
- Handle ethidium bromide with care, as it’s a DNA-intercalating agent, and dispose of ethidium bromide-containing solutions in dedicated containers.
- For single-cell experiments, quantification from microscope images are mandatory. However, if bulk measurements are desired, a fluorescence plate-reader can be used instead of a microscope.
- If higher concentrations of ethidium bromide are used, incubation times should be reduced. However, with very short incubation times differences in handling times will make the comparison of samples more difficult.
- The excitation and emission maxima of ethidium bromide are 524 nm and 605 nm, respectively. Hence, for ethidium bromide green excitation filters and orange emission filters should be used. Typically, standard microscope settings for Cy3/TRITC/rhodamine can be used, but for optimal performance the best combinations of available filters should be determined by the experimenter.
Recipes
- Culture medium for propagation of HelaCx26 or HelaCx43 cells
Mix 500 ml sterile DMEM with sterile 5 ml penicillin + streptomycin (100 U/ml penicillin and 100 µg/ml streptomycin final, optional) and 50 ml sterile FBS
Stored at 4 °C
- HBSS solution
Mix HBSS with 1M HEPES to final concentration of 20 mM (e.g. 1 ml 1 M HEPES for 50 ml final volume)
- HBSS solution devoid of Ca2+
Mix HBSS devoid of calcium and magnesium with 1 M HEPES to final concentration of 20 mM (e.g. 1 ml 1 M HEPES for 50 ml final volume), MgCl2 to final concentration of 1 mM (e.g. 50 µl from 1 M stock) and EGTA to final concentration of 0.1 mM (e.g. 10 µl from 0.5 M stock).
- HBSS stock with ethidium bromide
For 5 µM ethidium bromide final concentration use 2.82 µl at 0.7 mg/ml ethidium bromide stock solution in per ml HBSS
- Acid washed coverslips
Separate coverslips using tweezers and transfer them into a glass beaker containing an excess of a 1 M HCl solution. Make sure that the coverslips don’t stick to each other. Heat the coverslips to 50-60 °C for 4-16 h and carefully agitate from time to time. Discard the HCl solution and wash the coverslips extensively with distilled water (at least twice) followed by double-distilled water. Wash coverslips once with ethanol and store in ethanol at RT in a sealed container until use (e.g. a sterile 50 ml Falcon tube). Let the ethanol dry off each coverslip before you use it.
Acknowledgments
A.P. was a recipient of subsequent EMBO Long-Term and Marie Curie Intra-European Fellowships. This project was supported by ANR grant 2010 MIDI 007 01 to A.P. and P.J.S. and ERC Advanced Grant HOMEOEPITH to P.J.S. This protocol was adapted from “Retamal, M.A., Schalper, K.A., Shoji, K.F., Bennett, M.V., and Saez, J.C. (2007). Opening of connexin 43 hemichannels is increased by lowering intracellular redox potential. Proc. Natl Acad. Sci. USA 104, 8322-8327” and a first short version of the adapted protocol was published in Puhar et al. (2013).
References
- Evans, W. H., De Vuyst, E. and Leybaert, L. (2006). The gap junction cellular internet: connexin hemichannels enter the signalling limelight. Biochem J 397(1): 1-14.
- Paemeleire, K., Martin, P. E., Coleman, S. L., Fogarty, K. E., Carrington, W. A., Leybaert, L., Tuft, R. A., Evans, W. H. and Sanderson, M. J. (2000). Intercellular calcium waves in HeLa cells expressing GFP-labeled connexin 43, 32, or 26. Mol Biol Cell 11(5): 1815-1827.
- Puhar, A. and Sansonetti, P. J. (2014). Induction of connexin-hemichannel opening. Bio-protocol 4(17): e1220.
- Puhar, A., Tronchère, H., Payrastre, B., Tran Van Nhieu, G. and Sansonetti, P. J. (2013). A Shigella effector dampens inflammation by regulating epithelial release of danger signal ATP through production of the lipid mediator PtdIns5P. Immunity 39(6): 1121-1131.
- Tran Van Nhieu, G., Clair, C., Bruzzone, R., Mesnil, M., Sansonetti, P. and Combettes, L. (2003). Connexin-dependent inter-cellular communication increases invasion and dissemination of Shigella in epithelial cells. Nat Cell Biol 5(8): 720-726.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Puhar, A. and Sansonetti, P. J. (2014). Dye-uptake Experiment through Connexin Hemichannels. Bio-protocol 4(17): e1221. DOI: 10.21769/BioProtoc.1221.
Category
Cell Biology > Cell-based analysis > Transport
Cell Biology > Cell imaging > Fluorescence
Biochemistry > Protein > Structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











