- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
SGR-based Reporter to Assay Plant Transcription Factor-promoter Interactions
Published: Vol 4, Iss 16, Aug 20, 2014 DOI: 10.21769/BioProtoc.1214 Views: 10936
Reviewed by: Claudia CatalanottiKanika GeraAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Microplate-Based Expression Monitoring System for Arabidopsis NITRATE TRANSPORTER2.1 Using the Luciferase Reporter
Yoshiaki Ueda and Shuichi Yanagisawa
Dec 5, 2024 1784 Views

A Novel Gene Stacking Method in Plant Transformation Utilizing Split Selectable Markers
Guoliang Yuan [...] Xiaohan Yang
Feb 20, 2025 1980 Views

Assessing Metabolite Interactions With Chloroplastic Proteins via the PISA Assay
Anna Karlsson [...] Elton P. Hudson
May 5, 2025 2011 Views
Abstract
We developed an in vivo method to assay plant transcription factor (TF)–promoter interactions using the transient expression system in Nicotiana benthamiana (N. benthamiana) plants. The system uses the Arabidopsis stay green (SGR) gene as a reporter. Induction of SGR expression in N. benthamiana causes chlorophyll degradation and causes leaves to turn yellow.
Materials and Reagents
- Plant material
- 4 to 5 week old healthy N. benthamiana plants
- 4 to 5 week old healthy N. benthamiana plants
- Vectors and bacteria strains
- pDONR221 or other gateway DONR vector (Life Technologies, InvitrogenTM, catalog number: 12536-017 )
- Escherichia coli (E. coli) DH10B or similar cells for molecular cloning
- SGR reporter destination vector, SPDK 2388
Note: This vector contains the reporter gene SGR without promoter.
- Binary vector for TFs over-expression (e.g. LIC6 from Arabidopsis Biological Resource Center)
- Binary vector for expression of negative control protein [e.g. Actin7 (At5g09810) in LIC6]
- Agrobacterium tumefaciens (A. tumefaciens) strain GV2260
- pDONR221 or other gateway DONR vector (Life Technologies, InvitrogenTM, catalog number: 12536-017 )
- Other materials
Equipment
- Incubator (42 °C)
- 1 ml Tuberculin syringes without needle (Tyco, catalog number: 8881501400 )
- Controlled environment plant growth chamber (12 h of light per day, 21 °C)
Procedure
- Cloning of transcription factor of interest into SGR reporter vector
- Amplify promoter fragments (1-2 kb) from plant genomic DNA by high-fidelity PCR, and clone them into Gateway entry vector (pDONR221 or pDONR207) using BP reaction according to manufacturer’s instruction. This will generate the entry vector containing the promoter fragments.
- Transfer the promoter fragments into the destination vector SPDK2388 via LR reaction. The LR reaction should be set up as below:
100 ng entry vector containing promoters
100 ng destination vector SPDK2388
2 µl LR enzyme buffer
2 µl LR enzyme
ddH2O to 10 µl
Incubate at 25 °C overnight. Add 1 µl Proteinase K, incubate at 37 °C for 10 min. Transform the reaction mix into chemical competent cells of E. coli DH10B or similar cells, and select the transformants on 50 µg/ml spectinomycin (spectinomycin50) containing LB plates.
- Amplify promoter fragments (1-2 kb) from plant genomic DNA by high-fidelity PCR, and clone them into Gateway entry vector (pDONR221 or pDONR207) using BP reaction according to manufacturer’s instruction. This will generate the entry vector containing the promoter fragments.
- Prepare plants and Agrobacterium strains
- Grow N. benthamiana plants with 12 h day light, 22 °C in the growth chamber. At 4 to 5 weeks, the plants should have three to four large healthy leaves suitable for infiltration.
- Transform separately the destination vector SPDK2388 with the TF of interest and the control into A. tumefaciens GV2260 by using chemical or electro competent cells. Typically, 200-500 ng plasmids and 50 µl competent cells are used for the transformation. Plate all the transformed cells and select the transformants on spectinomycin100, streptomycin100, Rifampicin25, and Carbenicillin50 (SSRC) containing LB plates. More than 100 transformants should be expected.
- Grow N. benthamiana plants with 12 h day light, 22 °C in the growth chamber. At 4 to 5 weeks, the plants should have three to four large healthy leaves suitable for infiltration.
- Evaluate background expression of the promoter::SGR reporter constructs in N. benthamiana leaves
- Grow Agrobacterium GV2260 strains harboring the promoter::SGR construct overnight in ~20 ml LB media with SSRC.
- Spin down the Agrobacterium culture at 3,000 x g for 10 min, discard the supernatant, and suspend in infiltration media. Adjust the concentration of the culture to a series of OD600. For example, 0.05, 0.1, 0.3, and 0.6 OD. Incubate the culture at the room temperature for 3 h.
- Infiltrate N. benthamiana leaves with different concentration of Agrobacterium culture prepared in step C2 above. Make a small cut on the abaxial side of the leaves, use 1 ml syringes without needle to infiltrate Agrobacterium culture into the leaves. The culture should cover a spot of ~1 cm in diameter (see Figure 1). Infiltrate 1-2 spots for each OD concentration and infiltrate all samples with different ODs onto a single leaf. Repeat the infiltration onto a second leaf within the same plant.
- Keep infiltrated plants for 48 or 72 h in the same growth chamber and check the spots for signs of yellowing. Choose the highest OD concentration that shows no or slight yellowing for the following experiment.
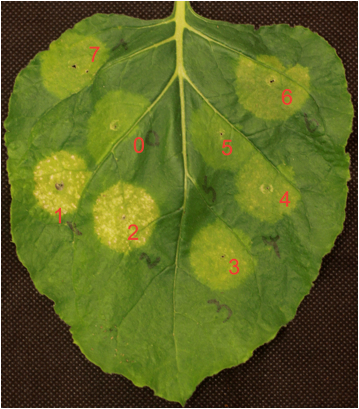
Figure 1. SGR reporter-based transcription factors and promoter interaction assay in Nicotiana benthamiana. The SUR1 promoter::SGR reporter construct was co-infiltrated with different MYB TFs (spots 1 to 7) or with a negative control Actin (spot 0). Only two MYBs (spot 1 and 2) showed severe yellowing signs indicating that these two MYBs activate SUR1 promoter.
- Grow Agrobacterium GV2260 strains harboring the promoter::SGR construct overnight in ~20 ml LB media with SSRC.
- Assay for TF-promoter interaction
- Grow separately Agrobacterium GV2260 strains harboring the promoter::SGR construct, TF construct, and actin control overnight in LB liquid media with SSRC.
Note: We have constructed an ATEC library in LIC6 binary vector for over-expression of 15,000 Arabidopsis genes, which include more than 700 transcription factors. All these clones are deposited to and can be ordered from ABRC. In ABRC, the ATEC clones are names as “DKL” plus AGI numbers. See Reference 3 for more details.
- Spin down the cultures min and re-suspend them in infiltration media. Adjust the OD of the reporter culture to the concentration determined in step C4. Adjust the OD of TF and actin control to 1.0.
- Mix Agrobacterium culture with the reporter and the TFs or control with equal volume (~ 1 ml each). Incubate at room temperature for 3 h.
- Infiltrate N. benthamiana leaves with the mixed cultures from above onto 1-2 spots of ~1 cm onto the same leaf. Control and treatment samples should be present on the same leaf to overcome leaf-to-leaf variability.
- Keep infiltrated plants for 48 to 96 h in the growth chamber. Record signs of yellowing of each spot at 24, 48, 72, and 96 h post infiltrations. The severity of yellowing indicates the strength of reporter gene expression by the corresponding TF. The spots containing TFs should be compared to the control spots to assess specificity of TF-promoter interactions. See example shown in Figure 1. Photograph the leaves to document the results.
- Further validation of the observed results could be performed using luciferase-based reporter assay as described in Ma et al. (2013).
- Grow separately Agrobacterium GV2260 strains harboring the promoter::SGR construct, TF construct, and actin control overnight in LB liquid media with SSRC.
Recipes
- Infiltration medium
Note: Prepare fresh media every time; sterilization is not required.
10 mM MgCl2
10 mM MES
200 µM acetosyringone
Acknowledgments
This protocol is adapted from the following paper: Ma et al. (2013). This work is supported by National Science Foundation grants DBI-0723722 and DBI-1042344, and UC Davis funds to SPDK.
References
- Hortensteiner, S. (2009). Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci 14(3): 155-162.
- Ma, S., Shah, S., Bohnert, H. J., Snyder, M. and Dinesh-Kumar, S. P. (2013). Incorporating motif analysis into gene co-expression networks reveals novel modular expression pattern and new signaling pathways. PLoS Genet 9(10): e1003840.
- Popescu, S. C., Popescu, G. V., Bachan, S., Zhang, Z., Seay, M., Gerstein, M., Snyder, M. and Dinesh-Kumar, S. P. (2007). Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc Natl Acad Sci U S A 104(11): 4730-4735.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ma, S. and Dinesh-Kumar, S. (2014). SGR-based Reporter to Assay Plant Transcription Factor-promoter Interactions. Bio-protocol 4(16): e1214. DOI: 10.21769/BioProtoc.1214.
Category
Plant Science > Plant biochemistry > Protein > Interaction
Plant Science > Plant molecular biology > DNA > Gene expression
Systems Biology > Interactome > Gene network
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









