- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
siRNA Screening for Genes Involved in HSV-1 Replication
Published: Vol 4, Iss 16, Aug 20, 2014 DOI: 10.21769/BioProtoc.1209 Views: 11243
Reviewed by: Pinchas TsukermanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Screening Method for CRISPR/Cas9 Inhibition of a Human DNA Virus: Herpes Simplex Virus
Werner M. Neuhausser [...] David M. Knipe
Sep 5, 2020 6728 Views

VirScan: High-throughput Profiling of Antiviral Antibody Epitopes
Ellen L. Shrock [...] Stephen J. Elledge
Jul 5, 2022 8583 Views

A Novel Method of Inducible Directed Evolution to Evolve Complex Phenotypes
Ibrahim S. Al’Abri [...] Nathan Crook
Oct 20, 2022 3062 Views
Abstract
Small interfering RNAs (siRNAs) are small (typically 18-24 nucleotides) RNA molecules capable of silencing gene expression post-transcriptionally and as such, they provide a simple method by which the role of individual genes in complex cellular systems can be easily assessed. As siRNAs are easy to use experimentally, and can be designed to target any gene (including pathogens), their use is perfectly suited to and easily adapted to high-throughput genome-wide screening methodologies and a range of phenotypic assays. Here we describe the use of a large siRNA library (>8,000 genes targeted individually) to screen for and identify host factors functionally involved in the replication of a human herpesvirus (Herpes simplex virus type 1; HSV-1) (Griffiths et al., 2013; Griffiths, 2013).
Materials and Reagents
- Hela cells (ATCC, catalog number: CCL-2 )
- siRNA library (e.g. human siGENOME SMARTpool Druggable Genome siRNA library 0.5 nmol) (Thermo Fisher Scientific, catalog number: G-004605-05 )
- 5x siRNA buffer (Thermo Fisher Scientific, catalog number: B-002000-UB-100 )
- RISC-free (RSCF) control siRNA (20 nmol) (Thermo Fisher Scientific, catalog number: D-001220-01-20 )
- Scrambled, non-targeting control siRNA (20 nmol, human) (Thermo Fisher Scientific, catalog number: D-001206-13-20 )
- Assay-specific control siRNAs
- Fetal bovine serum (FBS) (LabTech, catalog number: FCS-SA-10454 )
- Penicillin:streptomycin (5,000 units/ml each) (Lonza, catalog number: DE17-603E )
- 1x trypsin-EDTA liquid (0.05% Trypsin, 0.53 mM EDTA-4Na) (Life Technologies, catalog number: 25300096 )
- Phosphate buffered saline without magnesium or calcium (Lonza, catalog number: 17-516F )
- DMEM 4.5 g glucose/litre with L-glutamine and sodium pyruvate (Lonza, catalog number: BE12-604F )
- 1x DMEM/F12 1:1 (15 mM HEPES + L-glutamine, no phenol red) (Life Technologies, catalog number: 11039021 )
- Hank’s balanced salt solution (HBSS) (Thermo Fisher Scientific, catalog number: HYC-001-181W )
- Dharmafect 1 siRNA transfection reagent (Thermo Fisher Scientific, catalog number: T-2001 )
- Sterile DNase and RNase-free distilled water (Life Technologies, catalog number: 10977-049 )
- Ethanol (diluted in sterile RNase and DNase-free water)
- CellTiter-Blue® cell viability reagent (Promega Corporation, catalog number: G8081 )
- HSV-1-eGFP reporter virus (Arthur et al., 2001)
- Growth medium (see Recipes)
- Transfection medium (see Recipes)
Equipment
- Pipette tips for robotic liquid handler (Axygen Zymark 200 µl sterile tips) (VWR International, catalog number: AXYGZT-200-L-R-S )
- Polystyrene plate covers for 96-well plates (Thermo Fisher Scientific, catalog number: AB0752 )
- Aluminium heat-sealing film for storage microplates (Thermo Fisher Scientific, catalog number: AB-0559 )
- Adhesive plate seals (Starlab, catalog number: E2796-9793 )
- 384-well storage plates (Thermo Fisher Scientific, catalog number: AB-0781 )
- Black 384-well clear flat-bottomed, sterile, tissue culture-treated plates (VWR International, catalog number: 734-1640 )
- 75 cm2 filter cap tissue culture flasks (Sigma-Aldrich, catalog number: C7106 )
- 175 cm2 filter cap tissue culture flasks (Sigma-Aldrich, catalog number: C7481 )
- 50 ml centrifuge tube (Corning, product number: 430290 )
- Disposable hemocytometer (KOVA Glasstic slide 10 with counting grid) (HYCOR Biomedical, catalog number: 87144E )
- Class II Microbiological safety cabinet
- Humidified cell culture incubator (37 °C, 5% CO2)
- Pipette aid (Alpha laboratories, catalog number: 4-131-201-E )
- Centrifuge (e.g. Eppendorf, catalog number: 5811-000.010 )
- Light microscope
- 8-channel multichannel pipette (2-10 μl volume) (Thermo Fisher Scientific, catalog number: 4661000 )
- Multidrop 384 reagent dispenser (Thermo Fisher Scientific, catalog number: 5840150 )
- Multidrop 384 standard tube dispensing cassette (Thermo Fisher Scientific, catalog number: 24072670 )
- RapidPlate 384 robotic liquid handler (or equivalent) (discontinued) (QIAGEN)
- Microplate heat sealer (e.g. ALPS 50 V Microplate heat sealer) (Thermo Fisher Scientific, catalog number: AB-1443 )
- Gas-permeable adhesive microplate seals (Thermo Fisher Scientific, catalog number: AB-0718 )
- Barcode printer (e.g. Brady® LABXPERT™ printer) (Sigma-Aldrich, catalog number: Z664588 )
- Brady® LABXPERT™ printer labels (Thermo Fisher Scientific, catalog number: 12879858 )
- Fluorescent plate reader (e.g. POLARstar OPTIMA, BMG LABTECH)
- Stacker for plate reader (BMG LABTECH)
Software
- Microsoft Excel
Procedure
- Remove the lyophilised siRNA library (76 x 96-well plates) from storage at -80 °C and equilibrate to room temperature.
- Pellet well contents by centrifuging for 10 min at 1,000 rpm and replace plate seals with clean plastic lids.
- Clean a Multidrop 384 dispensing cassette with 3 x 25 ml 70% ethanol and rinse with 3 x 50 ml sterile DNase and RNase-free water.
- Prepare a sufficient volume of 1x siRNA buffer by diluting 5x siRNA buffer in sterile DNase and RNase-free water.
- Resuspend the 0.5 nmol siRNA library to 3 µM by dispensing 165 µl 1x siRNA buffer to columns 3-12 of the siRNA library plates (96-well plates) (see Note 1; Figure 1A).
- Using a label maker, print text or barcode labels for 384-well Master Plates (see Note 2).
- Adhere labels to 384-well storage plates (for a library of 76 x 96-well plates you will need 19 x 384-well storage plates and labels). These will be the 384-well Master Plates.
- Clean a dispensing cassette for the Multidrop 384, as above, and dispense 110 µl 1x siRNA buffer into columns 1 and 2 of the 384-well Master plate (see Note 3; Figure 1B).
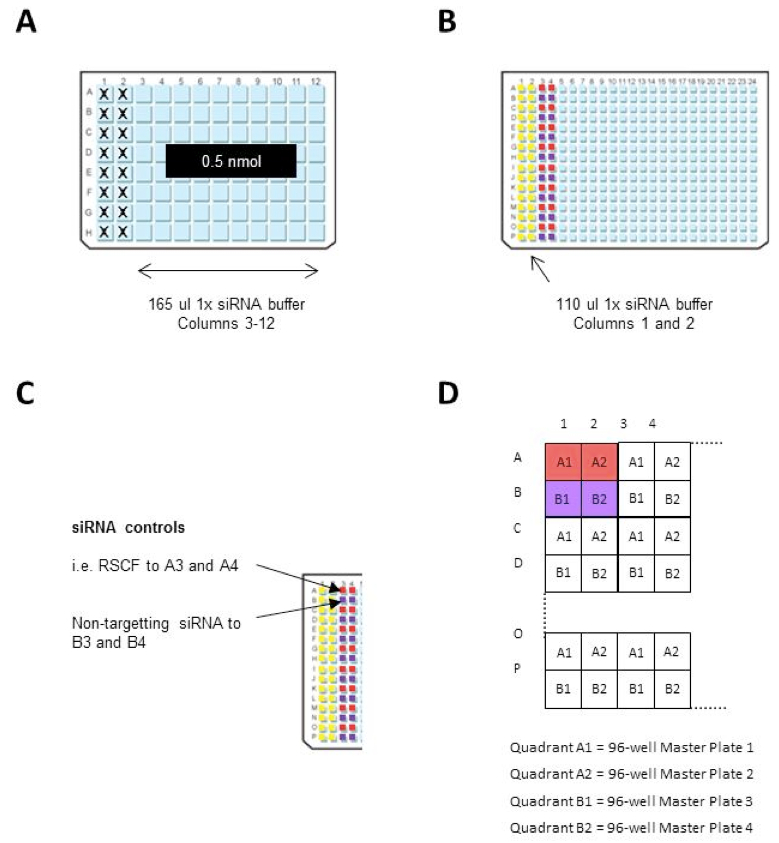
Figure 1. 96 and 384-well plate preparation - Dilute the assay-specific siRNA controls to 300 nM in 1x siRNA buffer (see Note 4).
- Manually transfer 110 µl of each siRNA control, in horizontal duplicates, to columns 3 and 4 in every 384-well Master Plate i.e. transfer RSCF siRNA to wells A3 and A4; transfer non-targeting siRNA to wells B3 and B4 etc. (Figure 1C).
- Dispense 100 µl 1x siRNA buffer to columns 5-24 in each 384-well master plate using a Multidrop 384.
- Using a robotic liquid handler (such as the QIAGEN RapidPlate 384), with an individual box of Zymark robotic tips for each siRNA 96-well Library Plate, transfer 11 µl from the siRNA library plates (3 µM siRNA) to the 384-well Master plate. Each 384-well plate takes 4 x 96-well siRNA Library plates (Figure 1D). These plates can be stored at -80 °C until needed.
- Use a microplate heat sealer and aluminium heat seals to seal the siRNA library plates before transferring to a -80 °C for long-term storage.
- Print appropriate barcode or text labels for the 384-well assay plates containing relevant information (replicate number, assay type etc.) (see Note 5).
- Stick labels to black, tissue-culture treated, flat, clear-bottomed 384-well plates.
- If frozen, thaw the 384-well Master Plates and briefly centrifuge for 1 min at 1,000 rpm before transferring 10 µl to three replicate assay plates using a robotic liquid handler and an individual box of Zymark robotic tips for each quadrant of the 384-well plate.
- Cover prepared assay plates with an adhesive plate seal, place in sealable bags and stored at -80 °C for up to 2 weeks before use (see Note 6).
- Calculate the number of low passage Hela cells in a confluent T175 flask, and the total number of cells required for your assay. Based on these numbers determine the number of 50% confluent T175 flasks required to provide the total number of cells for your assay.
- Several days before your assay, prepare Hela cells such that there will be sufficient 100% confluent T175 flasks the day before the siRNA transfection.
- The day before transfection, remove medium from the confluent T715 flasks of Hela cells, and rinse cells with warmed sterile phosphate buffered saline.
- Discard wash, and dislodge cells by incubating for around 5 min at 37 °C in 5 ml pre-warmed trypsin.
- Inactivate trypsin by adding 10 ml growth medium and transfer the pre-determined volume of cells to sufficient T175 flasks such that they will be 50% confluent the following day (see Note 7).
- The next day, thaw the replicate assay plates and centrifuge for 5 min at 1,000 rpm. Remove the plate seals and cover with lids.
- Dilute the Dharmafect 1 (DF1) transfection reagent to 0.6% in the appropriate volume of HBSS and add 10 µl to each well of the 384-well assay plates using a dispensing cassette for the Multidrop 384.
- Centrifuge the assay plates for 1 min at 1,000 rpm and incubate at room temperature for 20 min for transfection complexes to form.
- Remove growth medium from the Hela cells, rinse in PBS and dislodge cells by incubating for ~5 min at 37 °C in 5 ml trypsin.
- Inactivate the trypsin by adding 10 ml of transfection medium.
- Mix each flask well and pool together all cells before mixing well and removing 5 x 12 μl aliquots of cell suspension for counting in a disposable hemocytometer.
- Calculate the average cell density and dilute to 7.5 x 104 cells/ml.
- After the 20 min incubation time, transfer 40 µl cells to every well in the assay plates and incubate plate in a humidified incubator at 37 °C for 48 h to ensure gene depletion prior to investigating your assay output.
- After 48 h, determine the effect of gene depletion on cell viability.
- Thaw the CellTitre Blue reagent (or equivalent) and add 5 µl per well of each Cell Viability replicate plate using the Multidrop 384.
- Return the plates to the 37 °C incubator for 2 h before measuring the fluorescence signal (485 nm) of each plate using a POLARstar OPTIMA plate reader (or equivalent). The effect of siRNA depletion on cell viability is determined by comparing fluorescence to the average fluorescence of mock-transfected cells (columns 1 and 2) (see Note 8).
- To infect the duplicate assay plates with HSV-1-eGFP, thaw theHSV-1-eGFP virus stock in a 37 °C water bath until just thawed, before diluting to a multiplicity of infection (MOI) of 0.5 in phenol red-free DMEM: F12/15 mM HEPES/L-glutamine/5% FCS/pen-strep (see Note 9).
- Remove media from all plates by inverting and shaking over a suitable liquid waste container.
- Add 10 µl phenol red-free growth medium to columns 1 and 2 of every plate manually using a low volume (2-10 µl) 8-channel multichannel pipette (see Note 10).
- Add 10 µl diluted virus to the remaining wells (columns 3-24) using a Multidrop 384 and centrifuge for 2 min at 1,000 rpm to remove air bubbles and ensure the cell monolayer is covered by the small volume of inoculum.
- Transfer infection assay plates to the 37 °C incubator for 1 h to allow virus to adsorb before adding 50 µl phenol red-free growth medium to every well using the Multidrop 384.
- Cover the plates with a gas-permeable adhesive seal, stack and cover the top plate with a plastic lid before returning all assay plates to the incubator for 20 h.
- Place assay plates in the stacker equipment of a fluorescence plate reader and monitor virus replication as a measure of GFP fluorescence in a fluorescent plate reader every 3-4 h until replication plateaus (see Note 11).
- Once the virus replication cycle has completed, calculate the replication slope over the linear phase of growth for each well and determine the effect of gene depletion on virus replication by comparing the replication slope to the average replication slope of control wells (mock, RSCF and non-targeting siRNA-transfected) on a per plate basis. The LINEST equation in Microsfot Excel can be used to calculate replication slopes. This formula utilises the ‘known x’ values (the hours post-infection) and the ‘known y’ values (the relative fluorescence at each time post-infection). Firstly, generate a table with the ‘Hours post-infection’ and the relative fluorescence of your samples (Figure 1A). Plot these data and identify the linear phase of replication (Figure 1B; red line indicates linear phase). Use the linest formula to calculate the replication slope, and normalise the slope of each condition to that of your control 9in this example, normalise to ‘Mock’ transfected cells; Figure 1C-D).
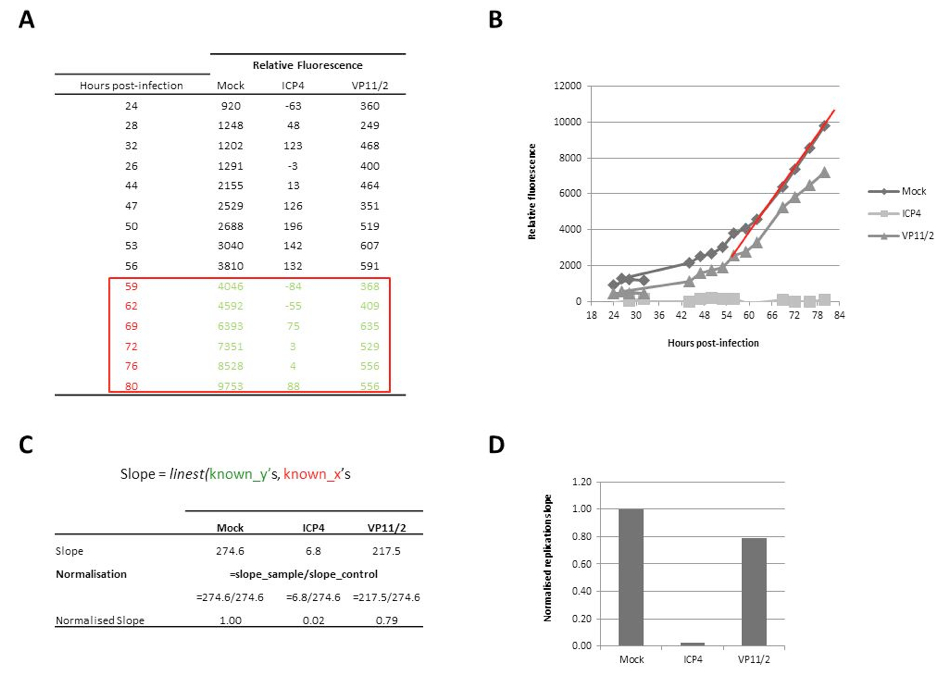
Figure 2. Calculation of replication slopes
Notes
- Library plates are diluted 1: 10 to 300 nM when generating the 384-well Master Plates, and diluted a further 1: 6 during the assay. This gives a final siRNA concentration of 50 nM. If a higher/lower siRNA concentration is preferred, adjust the resuspension volumes accordingly. columns 1 and 2 of the 96-well siRNA Library plates are empty to enable the user to include assay-specific controls.
- As siRNAs are temperature sensitive, we recommend the generation of a 384-well master plate which contains sufficient siRNA to complete three full screens in triplicates. siRNA can be dispensed into replicate assay plates at the same time to ensure an equal number of freeze-thaw cycles of assay plates and reproducible assay results.
- The addition of siRNA buffer alone to columns 1 and 2 of each 384-well master plate will result in untransfected control cells. For our virus replication assay, these will remain uninfected.
- Be sure to include mock-transfected cells (transfection reagent alone), siRNA controls (such as RISC-free and a scrambled, non-targeting siRNA) and assay-specific siRNAs which display a known range of phenotypes (strong positive effect, moderate effect, strong negative effect).
- For a virus replication assay we recommend carrying out each assay in triplicate. One replicate should be used for a cell viability assay (as virus will not replicate if siRNA gene depletion has a cytotoxic effect) and the virus replication assay is done in duplicates.
- Whilst diluted siRNA will be stable for some time at -80 °C the risk of evaporation of such a small volume is high. We would recommend storing assay plates for no longer than 2 weeks before use.
- siRNA transfection and virus replication is optimal when cells were sub-confluent (~50%) and freshly passaged prior to use.
- The Cell Titre Blue reagent is metabolised to produce a fluorescent signal. The cell viability is therefore demonstrated by the rate of metabolism of the cells. The effect of gene depletion on cell viability is determined by comparing the fluorescence of siRNA-transfected wells to the average fluorescence of control, non-transfected wells (columns 1 and 2) on a per plate basis to avoid inter-plate variations. Normalising the fluorescence in this way enables identification of cytotoxic siRNAs as well as those increase cell proliferation. As per manufacturer’s instructions, a fluorescence value of <70% of the control cells is considered cytotoxic. Cytotoxic siRNAs are excluded from subsequent data analyses.
- It is important to infect cells with a known quantity of virus (multiplicity of infection; MOI) to generate a suitable growth curve. We find a multiplicity of infection (MOI) of 0.5 [0.5 virus plaque forming units (PFU) per cell] gives an ideal kinetic growth curve from 20 to 80 h post-infection. The MOI is dependent upon the cell number and volume used for the infection, so the appropriate dilution factor should be calculated based upon the starting titre of your virus stock. For example, to infect 3 x 103 cells at MOI 0.5 in 10 μl we need to have a virus preparation containing 1.5 x 105 PFU/ml. A starting stock of 6 x 107 PFU/ml should therefore be diluted 1 in 400.
- Tissue culture plasticware, cells and growth medium all have some level of fluorescence. When utilising fluorescent reporter genes it is therefore essential to have appropriate controls to provide background fluorescence readings. For this assay, the fluorescence from uninfected cells is used as background.
- Replication is monitored over regular intervals to enable a complete growth curve (fluorescence over time) to be plotted. When comparing HSV replication between untreated and treated samples we use the slope of replication over the linear growth phase. It is therefore important to have as many measurements over this phase as possible (with a minimum of 6 for statistical reliability of the slope calculation). A test replication assay with your own equipment will allow the time of linear growth to be established, and assay timings can be adjusted to ensure measurements can be taken over this period.
Recipes
- Growth medium
DMEM
5% FCS
1% pen-strep - Transfection medium
Phenol red-free DMEM: F12
15 mM HEPES
L-glutamine
5 % FCS
Acknowledgments
This protocol has been edited from that described in Griffiths et al. (2013) PLOS Pathogens. The authors are grateful to Prof. Peter Ghazal for provision of the siRNA libraries, and Marie Craigon for technical assistance with library handling, management and establishment of protocols. The authors are grateful to the MRC (G0501453 J.H.), BBSRC (BB/B019621/1 P.G.), Scottish Enterprise (P.G.) and the Wellcome Trust (WT066784 P.G.) for funding.
References
- Arthur, J. L., Scarpini, C. G., Connor, V., Lachmann, R. H., Tolkovsky, A. M. and Efstathiou, S. (2001). Herpes simplex virus type 1 promoter activity during latency establishment, maintenance, and reactivation in primary dorsal root neurons in vitro. J Virol 75(8): 3885-3895.
- Griffiths, S. J., Koegl, M., Boutell, C., Zenner, H. L., Crump, C. M., Pica, F., Gonzalez, O., Friedel, C. C., Barry, G., Martin, K., Craigon, M. H., Chen, R., Kaza, L. N., Fossum, E., Fazakerley, J. K., Efstathiou, S., Volpi, A., Zimmer, R., Ghazal, P. and Haas, J. (2013). A systematic analysis of host factors reveals a Med23-interferon-lambda regulatory axis against herpes simplex virus type 1 replication. PLoS Pathog 9(8): e1003514.
- Griffiths, S. J. (2013). Screening for host proteins with pro- and anti-viral activity using high-throughput RNAi in ‘Virus-Host Interactions’. In: Methods in Molecular Biology. Vol. 1064: 71-90.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Griffiths, S. J. and Haas, J. (2014). siRNA Screening for Genes Involved in HSV-1 Replication. Bio-protocol 4(16): e1209. DOI: 10.21769/BioProtoc.1209.
- Griffiths, S. J., Koegl, M., Boutell, C., Zenner, H. L., Crump, C. M., Pica, F., Gonzalez, O., Friedel, C. C., Barry, G., Martin, K., Craigon, M. H., Chen, R., Kaza, L. N., Fossum, E., Fazakerley, J. K., Efstathiou, S., Volpi, A., Zimmer, R., Ghazal, P. and Haas, J. (2013). A systematic analysis of host factors reveals a Med23-interferon-lambda regulatory axis against herpes simplex virus type 1 replication. PLoS Pathog 9(8): e1003514.
Category
Systems Biology > Genomics > Sequencing
Molecular Biology > RNA > RNA interference
Molecular Biology > DNA > Mutagenesis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










