- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro DNA Polymerization Activity Assay Using Cell-free Extracts
Published: Vol 4, Iss 16, Aug 20, 2014 DOI: 10.21769/BioProtoc.1207 Views: 9249
Reviewed by: Fanglian HeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Introduction and Sequencing of Patient-isolated HBV RT Sequences into the HBV 1.2-mer Replicon
Sung Hyun Ahn [...] Kyun-Hwan Kim
Apr 20, 2015 9696 Views

Estimation of the Minimum Number of Replication Origins Per Chromosome in any Organism
Marcelo S. da Silva
Oct 20, 2020 4214 Views

Protocol to Mine Unknown Flanking DNA Using PER-PCR for Genome Walking
Zhou Yu [...] Haixing Li
Feb 20, 2025 1508 Views
Abstract
This protocol has been designed to measure the in-vitro DNA polymerization activity in crude cell extracts of the Antarctic bacterium Pseudomonas syrinagae Lz4W. This bacterium can grow at 4 °C with optimum growth rate at 22 °C. The slow growth rate of the bacterium observed at low temperature (4 °C) compared to higher temperature (22 °C) can be attributed to the reduced rate of DNA replication at low temperature. Here we describe a protocol which we have used to quantify the in vitro DNA polymerization of cell extracts at two different temperatures.
Keywords: DNA synthesisMaterials and Reagents
- Pseudomonas syringae (P. syringae) Lz4W strain (Shivaji et al., 1989)
- Escherichia coli (E. coli) DH5alpha strain
- Random primer labelling kit (JONAKI Laboratory, catalog number: LCK 2 )
Note: Many companies like Roche Diagnostics, Thermo Fisher Scientific, Agilent, Clontech etc. provide similar kit.
- Wizard Genomic DNA Purification Kit (Promega Corporation, catalog number: A1120 )
- (Alpha 32P)-dATP (JONAKI Laboratory, catalog number: LCP 103 )
- Lysozyme (Roche Diagnostics, catalog number: 10153516103 )
- 10% trichloroacetic acid (TCA) (Sigma-Aldrich, catalog number: T6399 )
- KCl (Sigma-Aldrich, catalog number: P9541 )
- 50 mM Tris.HCl (pH 7.5)
- 10% sucrose (Sigma-Aldrich, catalog number: 84097 )
- Peptone (HiMedia Laboratories, catalog number: RM 001-500G )
- Yeast extracts (HiMedia Laboratories, catalog number: RM 027-500G )
- Stop buffer (see Recipes)
- Lysozyme solution (see Recipes)
- ABM broth (Pavankumar et al., 2010) (see Recipes)
Equipment
- BD PrecisionGlide needles 22G (BD, catalog number: 305156 )
- Liquid scintillation analyzer (PerkinElmer, model: Tri-Carb 2900 TR )
- Centrifuge (Eppendorf, model: 5810R, catalog number: 5811000.010 )
- NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, catalog number 8482 )
- Micro centrifuge tubes (1.5 ml) (tarsons, catalog number: 500010 )
- F12- ED refrigerated/heating circulator (JULABO GmbH, catalog number: 9116612 )
- Plastic scintillation vials (PerkinElmer, catalog number: 6000477 )
- Separate laboratory design and workspace for radioactive work with proper guidelines
Procedure
- Preparation of cell extracts
The basic method of cell extracts preparation was adapted from methods described earlier (Wickner et al., 1972; Villani et al., 1978).- Inoculate P. syringae Lz4W strain from a single colony (streaked on ABM agar plate and was incubated at 22 °C for 24-36 h) in 5 ml ABM liquid media (autoclaved in 25 ml flask). Grow it for overnight at 22 °C shaking at 200 rpm.
- Inoculate a fresh secondary culture using the overnight grown cultures inoculum in a new 20 ml ABM broth (autoclaved in 100 ml flask) in 1:100 ratio dilution and grow at 22 °C till exponential growth phase (OD600~ 0.3 to 0.4, 6-7 h growth).
- Pellet down cells by centrifuging at 10,000 rpm for 5 min at 22 °C.
- Discard the supernatant, wash the pellet with 1 ml of buffer containing 10% sucrose containing 50 mM Tris.HCl (pH 7.5). Resuspend the pellet in 0.002 volume (40 µl for 20 ml culture) of 10% sucrose containing 50 mM Tris.HCl (pH 7.5) at room temperature (22-25 °C).
- Add 1/10 th volume (4 µl) of a lysozyme solution and 0.025 volume (1 µl) of 4 M KCl. Incubate the mixture at 0 °C (on ice) for 30 min.
- Centrifuge the lysate at 14,000 rpm for 45 min at 2 °C.
- Supernatant was transferred to a fresh microcentrifuge tube and stored at 0 °C (on ice). Quantify the total protein concentration using NanoDrop 1000 spectrophotometer.
Note: Although this cell lysate is stable on ice for few hours, in most of our experiments we used this freshly prepared lysate within 30-60 min of preparation.
- Inoculate P. syringae Lz4W strain from a single colony (streaked on ABM agar plate and was incubated at 22 °C for 24-36 h) in 5 ml ABM liquid media (autoclaved in 25 ml flask). Grow it for overnight at 22 °C shaking at 200 rpm.
- Preparation of template DNA (E. coli genomic DNA)
- Isolate genomic DNA from E. coli strain MC4100 using genomic DNA isolation kit.
Note: This step to be done one day before preparing the Pseudomonas cell extracts.
- Shear the genomic DNA by passing it through a large gauge (22G) needle for several times (15-16 times to get 6-10 kb size of sheared DNA).
- Determine the concentration of DNA by taking readings in NanoDrop 1000 spectrophotometer.
- Denature the template DNA by incubating it for 2-3 min in a boiling water bath, and then immediately freeze it into ice water bath. This DNA will be used as a template to assay the DNA polymerization activity using any Gram-negative bacterial cell extracts.
- Isolate genomic DNA from E. coli strain MC4100 using genomic DNA isolation kit.
- DNA polymerization activity assay
- Pre-incubate the aliquots of Pseudomonas cell lysate at the desired temperatures for 10 min. We used 22 °C and 4 °C for P. syringae cell extracts.
- Prepare replicas (2 to 3) of two assay mixtures (experimental): One set to be incubated at 22 °C and another set at 4 °C.
- Each assay mixture (50 µl) will have 1 μg of heat denatured template DNA (sheared E. coli genomic DNA), Random primer buffer (1x), Random hexanucleotide primers (1x), 0.8 mM each of dCTP, dGTP, dTTP and 40 μCi of [alpha-32P]-dATP. Pre-incubate the samples at the experimental temperatures (22 °C and 4 °C) for at least 5 min.
Note: Reaction volume to be adjusted using sterile Milli-Q water, considering the volume of cell extracts to be added, to make the final volume of 50 µl.
- Prepare two more sets of assay mixtures as controls and process them as above. Control (negative) assay mixtures will have everything except the template DNA.
- In the mean time, label the tubes for collecting reaction samples at different time points. For example, for six time points, take 6 micro centrifuge tubes for each assay mixture and labeled them as 0, 15, 30, 60, 90 and 120 sec. Add 30 µl of E. coli unlabeled denatured DNA (50 µg) in stop buffer and 40 µl of 10% TCA into each tube as stop buffer.
Notes:- Addition of E. coli unlabeled denatured DNA (cold DNA) increases the precipitation efficiency of labeled DNA (hot DNA).
- The cold DNA was isolated from E. coli grown cells, heat denatured and immediately transferred in ice water.
- Addition of E. coli unlabeled denatured DNA (cold DNA) increases the precipitation efficiency of labeled DNA (hot DNA).
- Now, to start the experiment, add 100 µg of pre-incubated cell lysates into the assay mixtures (step C4) which were incubated at 22 °C and 4 °C.
- Remove 10 µl of aliquote from each assay tubes at different time intervals (0, 15, 30, 60, 90 and 120 sec) and stop the reaction by adding into corresponding labeled tubes.
- Incubate these tubes for 1 h at 0 °C to precipitate the labeled DNA and then centrifuge at 14,000 rpm for 15 min at 4 °C.
- Remove the supernatant from the tubes containing unincorporated labels. To remove the drags, invert the tubes onto tissue paper or blotting paper for 2-3 min.
- Place centrifuge tubes into plastic scintillation vials and measure the radioactivity directly without adding any scintillation fluid (cherenkov counting) in a scintillation counter.
- Subtract the values (CPM) of control from the experimental value and calculate the rate of radioactivity incorporation.
- The radioactivity (count per minute or CPM) incorporation into DNA or polymerization rate can be expressed as CPM per minute per milligram of protein in the cell extracts.
- Pre-incubate the aliquots of Pseudomonas cell lysate at the desired temperatures for 10 min. We used 22 °C and 4 °C for P. syringae cell extracts.
Representative data
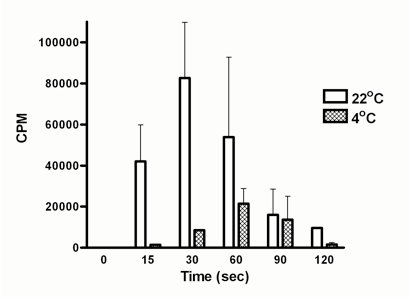
Figure 1. DNA polymerization activity is higher at 22 °C compared to 4 °C. DNA polymerization activity was assessed using Pseudomonas cell extracts as described above. Incorporation of radioactive nucleotides was observed immediately after addition of cell lysate both at 22 °C and 4 °C. Activity reduced after 30 sec at 22 °C whereas in case of 4 °C reaction it went down after 60 sec. This decrease in radioactivity count could be due to higher activity of exonucleases present in the cell extracts. Therefore, polymerization rate should be calculated from the values obtained from the linear curves generated within 60 sec of experiment under the conditions. The maximum CPM achieved by cell lysate was used to calculate polymerization rate as CPM per minute.
Recipes
- Stop buffer
10 mM Tris.HCl (pH 7.5)
100 mM EDTA
- Lysozyme solution
2 mg/ml in 0.025 M Tris.HCl (pH 7.6)
- ABM media
0.5% peptone
0.2% yeast extracts
Acknowledgments
The basic method of cell extracts preparation was adapted from methods described earlier (Wickner et al., 1972; Villani et al., 1978). The work was financed by a Grant from the Department of Science and Technology (DST), and by the Council of Scientific and Industrial Research (CSIR), Government of India.
References
- Pavankumar, T. L., Sinha, A. K. and Ray, M. K. (2010). All three subunits of RecBCD enzyme are essential for DNA repair and low-temperature growth in the Antarctic Pseudomonas syringae Lz4W. PloS One 5(2): e9412.
- Sinha, A. K., Pavankumar, T. L., Kamisetty, S., Mittal, P. and Ray, M. K. (2013). Replication arrest is a major threat to growth at low temperature in Antarctic Pseudomonas syringae Lz4W. Mol Microbiol 89(4): 792-810.
- Shivaji, S., Rao, N. S., Saisree, L., Sheth, V., Reddy, G. S. and Bhargava, P. M. (1989). Isolation and identification of Pseudomonas spp. from Schirmacher Oasis, Antarctica. Appl Environ Microbiol 55(3): 767-770.
- Villani, G., Boiteux, S. and Radman, M. (1978). Mechanism of ultraviolet-induced mutagenesis: extent and fidelity of in vitro DNA synthesis on irradiated templates. Proc Natl Acad Sci U S A 75(7): 3037-3041.
- Wickner, R. B., Wright, M., Wickner, S. and Hurwitz, J. (1972). Conversion of phiX174 and fd single-stranded DNA to replicative forms in extracts of Escherichia coli. Proc Natl Acad Sci U S A 69(11): 3233-3237.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sinha, A. K. and Ray, M. K. (2014). In vitro DNA Polymerization Activity Assay Using Cell-free Extracts. Bio-protocol 4(16): e1207. DOI: 10.21769/BioProtoc.1207.
Category
Microbiology > Microbial genetics > DNA > DNA replication
Molecular Biology > DNA > DNA synthesis
Molecular Biology > DNA > DNA labeling
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









