- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
DNA Fragmentation Analysis
Published: Vol 4, Iss 15, Aug 5, 2014 DOI: 10.21769/BioProtoc.1203 Views: 18154
Reviewed by: Fang XuZhaohui LiuRu Zhang

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Efficient Transient Gene Knock-down in Tobacco Plants Using Carbon Nanocarriers
Gozde S. Demirer and Markita P. Landry
Jan 5, 2021 6382 Views

Faster Bacterial Gene Cloning Using the Brick into the Gateway (BiG) Protocol
Flaviani G. Pierdoná [...] Fabio T. S. Nogueira
Dec 20, 2022 2363 Views
Abstract
DNA fragmentation with length corresponding to multiple integer of approximately 180 base pairs is a distinct feature of apoptosis in animals and programmed cell death in plants. This feature can simply be detected by DNA gel electrophoresis followed by ethidium bromide staining, although in some cases it is difficult to distinguish the DNA laddering. We herein describe a protocol to detect a programmed cell death-associated DNA laddering of plant tissues. After agarose-gel electrophoresis of genomic DNA, Southern hybridization using DIG-labeled genomic DNA probe is performed, that improves detection of DNA laddering.
Keywords: Programmed cell deathMaterials and Reagents
- Fresh plant tissues
- Nucleon PhytoPure DNA extraction kits (General Electric Company, catalog number: RPN8510 )
- GeneMate LE Agarose (BioExpress, catalog number: E-3120 )
- Biodyne Plus Nylon Membrane (Pall, catalog number: 60406 )
- Wizard DNA Clean-Up System (Promega, catalog number: A7280 )
- DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics, catalog number: 11585614910 )
- X-ray film
- Saline sodium citrate (SSC)
- Depurination solution (see Recipes)
- Denaturation solution (see Recipes)
- Neutralization solution (see Recipes)
- 1x TAE buffer (see Recipes)
Equipment
- Plastic container for gel and membrane treatment
- Centrifuge (Eppendorf, model: 5415R )
- Spectrophotometer (Shimadzu, model: BioSpec-nano )
- Shaker (BIO CRAFT, model: BC-700 )
- Vacuum transfer apparatus (BIO CRAFT, model: BS-31 )
- Hybridization oven and UV crosslinker (UVP, model: HybriLinker HL-2000 )
- Vacuum concentrator (SavantTM, model: DNA SpeedVac DNA110 )
- Hybridization incubator (TAITEC, model: HB-80 )
Procedure
- Grind plant tissues in liquid nitrogen and extract genomic DNA using PhytoPure according to user manual.
Notes:- Isopropanol precipitation is done at -20 °C for at least 30 min.
- Other plant genomic DNA isolation procedures are acceptable.
- Isopropanol precipitation is done at -20 °C for at least 30 min.
- Quantity and quality of the isolated genomic DNAs are checked by spectrophotometer and/or agarose gel electrophoresis.
Note: We used DNAs with OD260/280 > 1.8.
- 1-5 μg of genomic DNAs are loaded to a 2% agarose gel.
Note: 100-bp ladder marker is also loaded to estimate the molecular weight of fragmented DNA.
Note: Smaller amount (< 1 μg) of DNA may be possible to detect, although we have never tried it.
- Electrophoresis at 50 volt for 2 h in 1x TAE buffer.
Note: We used an agarose gel of 110 x 100 x 6 mm.
- Ethidium bromide staining and exposure to ultraviolet light.
Note: Check the positions of the bands of molecular marker in the gel.
- Place the agarose gel into a plastic container in depurination solution (250 ml) and gently agitate on a shaker for 10 min (or until bromophenol blue turned yellow).
Note: The depurination solution partially hydrolyzes large DNA fragments to help sufficient transfer of DNA fragments.
- Rinse the gel by dH2O for three times.
- Treat the gel in denaturation solution (250 ml) for 15 min twice.
Note: The denaturation solution denatures double-stranded DNA for efficient hybridization.
- Rinse the gel by dH2O for three times.
- Treat the gel in neutralization solution (250 ml) for 15 min twice.
Note: The neutralization solution neutralizes pH of the gel.
- Place the gel on a vacuum transfer apparatus with Biodyne Plus membrane presoaked in 20x SSC buffer and perform transfer for 2 h according to the manufacturer’s instructions.
Note: Filter blotting using 20x SSC is optional.
- Fix the DNA to the membrane by UV crosslinking and wash the membrane with sterilized dH2O.
Notes:- The membrane is wrapped in Saran Wrap prior to exposure to UV.
- UV energy dosage is 120 mJ/cm2.
- The membrane is wrapped in Saran Wrap prior to exposure to UV.
- Dry the membrane.
Note: This dried membrane can be stored in wrap at 4 °C.
- Preparation of DIG-labeled probe.
- Digest 2 μg of genomic DNA by 50 units of restriction enzyme at 37 °C for 12 h.
Note: 4-base cutters (e.g. HaeIII, MspI) are suitable.
- Purify the DNA using Wizard DNA Clean-Up System.
Note: Phenol-chloroform extraction and ethanol precipitation is optional.
- Dry the purified DNA solution using vacuum concentrator.
- Resolve the DNA with 16 μl of sterilized dH2O.
- Boil the DNA sample for 10 min and chill on ice to maintain the denatured single-strand DNA.
- Add 4 μl of DIG-High Prime (5x conc.) and mix.
- Incubate for 20 h at 37 °C.
- Stop the reaction by heating to 65 °C for 10 min.
- The reaction mixture can be stored at -20 °C.
- Digest 2 μg of genomic DNA by 50 units of restriction enzyme at 37 °C for 12 h.
- Place the membrane in a hybridization bottle and perform pre-hybridization for at least one hour and hybridization over-night using DIG Easy Hyb at 44 °C in a hybridization oven.
Note: In the case of the probe above mentioned, 2-3 μl of probe is added to the 5 ml hybridization buffer.
- Wash the membrane into a plastic container with 1x SSC and 0.2% SDS at 68 °C in a hybridization incubator for 15 min twice.
Note: Shake the container as fast as possible, making sure that the membrane keeps moving, and the solution should be changed every time.
- Wash the membrane into a plastic container with 0.1x SSC and 0.5% SDS at 68 °C in a hybridization incubator for 20 min twice.
- Perform detection according to the DIG-High Prime DNA Labeling and Detection Starter Kit II user manual.
- Expose chemiluminescent signal to X-ray film or to an imager. DNA laddering is visualized (Figure 1).
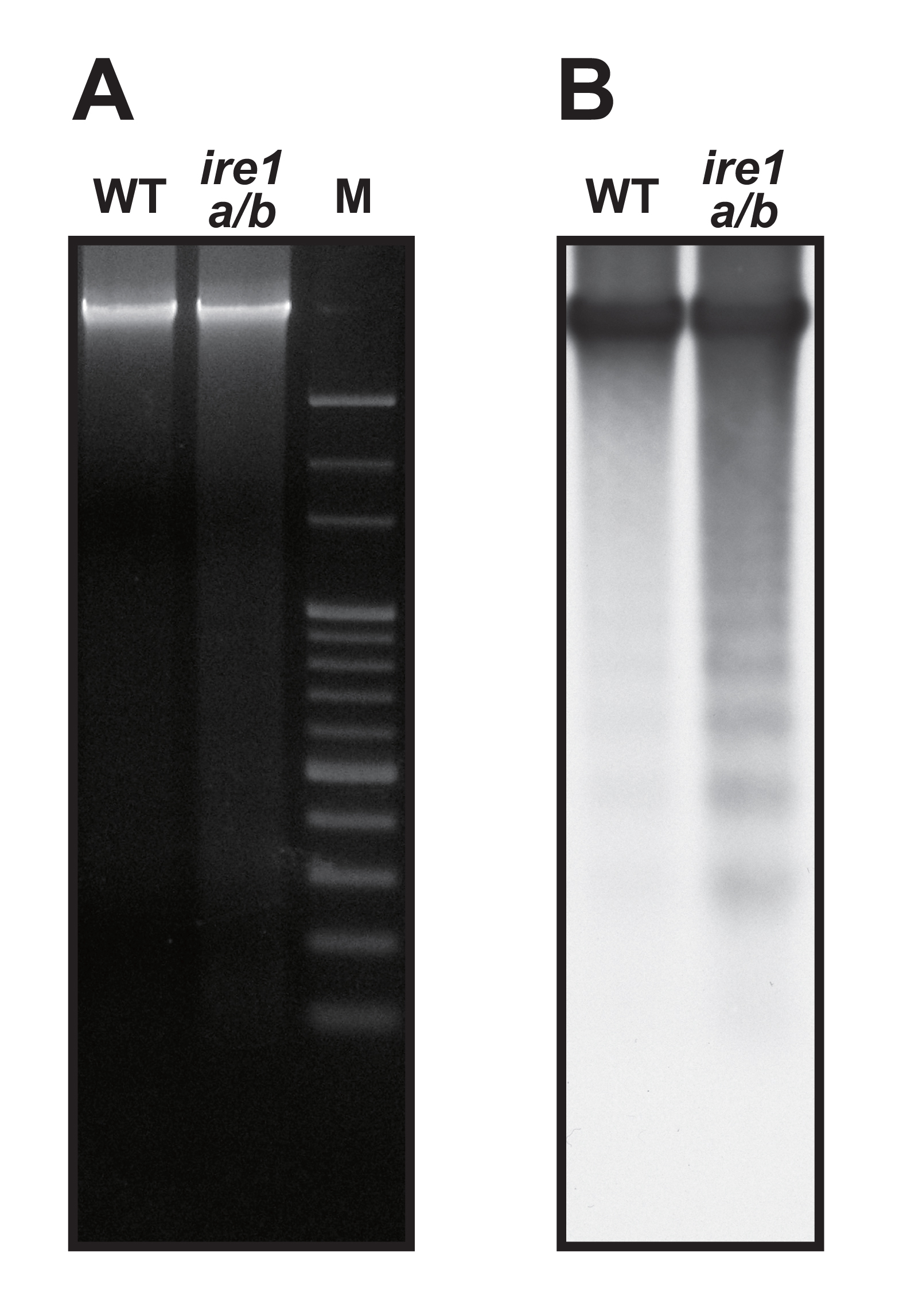
Figure 1. Detection of DNA laddering. Seedlings of Arabidopsis wild-type (WT) and ire1a/b mutant were treated with tunicamycin (5 µg/ml) for 48 h. (A) Ethidium bromide staining of genomic DNAs extracted form aerial parts of seedlings. M, 100-bp ladder marker. (B) Southern blot of the agarose gel in (A) hybridized with DIG-labeled HaeIII/MspI-digested Arabidopsis genomic DNA probe.
Recipes
- Depurination solution (500 ml)
Mix 25 ml of 5 N HCl (0.25 N) with 475 ml of dH2O
Stored at room temperature
- Denaturation solution (500 ml)
10 g of NaOH (0.5 N)
43.83 g of NaCl (1.5 M)
Add dH2O to 500 ml
Stored at room temperature
- Neutralization solution (500 ml)
30.3 g of Tris (0.5 M)
87.66 g of NaCl (3 N)
Add dH2O to 450 ml
Adjust pH to 7.5 by conc. HCl
Add dH2O to 500 ml
Stored at room temperature
- 1x TAE buffer
40 mM Tris-acetate
1 mM EDTA
Acknowledgments
This protocol was adapted from Mishiba et al. (2013).
References
- Mishiba, K., Nagashima, Y., Suzuki, E., Hayashi, N., Ogata, Y., Shimada, Y. and Koizumi, N. (2013). Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc Natl Acad Sci U S A 110(14): 5713-5718.
- Wyllie, A. H. (1980). Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature 284(5756): 555-556.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mishiba, K., Nagashima, Y., Hayashi, N. and Koizumi, N. (2014). DNA Fragmentation Analysis. Bio-protocol 4(15): e1203. DOI: 10.21769/BioProtoc.1203.
Category
Plant Science > Plant molecular biology > DNA > DNA modification
Molecular Biology > DNA > DNA damage and repair
Molecular Biology > DNA > Electrophoresis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










