- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantifying Fruit Dehiscence Using the Random Impact Test (RIT)
Published: Vol 4, Iss 15, Aug 5, 2014 DOI: 10.21769/BioProtoc.1200 Views: 9130
Reviewed by: Tie Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Plate Growth Assay to Quantify Embryonic Root Development of Zea mays
Jason T. Roberts [...] David M. Braun
Oct 20, 2023 2256 Views

Detection and Quantification of Programmed Cell Death in Chlamydomonas reinhardtii: The Example of S-Nitrosoglutathione
Lou Lambert and Antoine Danon
Aug 5, 2024 1589 Views

Direct Plant Regeneration From Immature Male Inflorescence of Banana (Musa spp.)
Pradeep Chand Deo
Oct 20, 2025 1438 Views
Abstract
Fruit dehiscence is an important evolutionary and agronomic trait. For quantifying and comparing the exact fruit dehiscence capability between individual plants, the random impact test has been described (Morgan et al., 1998; Bruce et al., 2002; Arnaud et al., 2010). Here, we describe the random impact test optimized to measure dehiscence capability in the Brassicaceae plant Lepidium campestre (L. campestre). However, with slight alterations regarding agitation force, agitation time, and drying conditions, the test should be applicable to a wide range of plant species with dehiscent fruits.
Keywords: Random impact testMaterials and Reagents
- L. campestre plants
Equipment
- An open container (e.g. Falcon tube)
- Desiccator or climate chamber allowing for the control of temperature and relative humidity
- A number of steel balls (in our case with a diameter of 5 mm)
- Mixer Mill (MM 400/Retsch) including two grinding jars (Figure 1)

Figure 1. Mixer Mill (MM400/Retsch)
- A pair of tweezers
Software
- Microsoft Excel
Procedure
- Grow L. campestre plants until fruits are yellow and completely dry.
- Carefully harvest at least 60 fruits per plant without manually causing fruits to open.
- Place the fruits in an open container (e.g. Falcon tube) and keep them under constant environmental conditions (25 °C and 50% relative humidity) for at least 3 days to achieve consistent moisture content.
- Count 20 fruits and place them in a grinding jar of the mixer mill together with six of the 5-mm steel balls (Figure 2).

Figure 2. 20 ripe fruits of L. campestre (left) placed in a grinding jar of the mixer mill together with six 5-mm steel balls (right)
- Carefully close the grinding jar, fasten it to the mixer mill and agitate for 5 sec at 9 hz.
- Remove the fruits from the grinding jar and determine the number of open fruits by counting the number of fully intact fruits (Figure 3).

Figure 3. Fully intact fruits are counted and open fruits are removed
- Put the intact fruits back to the grinding jar and agitate at 9 hz for another 5 sec, then count again.
- Repeat these two steps (agitating and counting) another 4 times (or until no intact fruits are left) with agitation times of 10, 20, 40, and 80 sec (overall resulting in cumulative times of 5, 10, 20, 40, 80, and 160 sec).
- Plot the time (sec) against the number of open fruits to end up with a graph like shown in Figure 4.
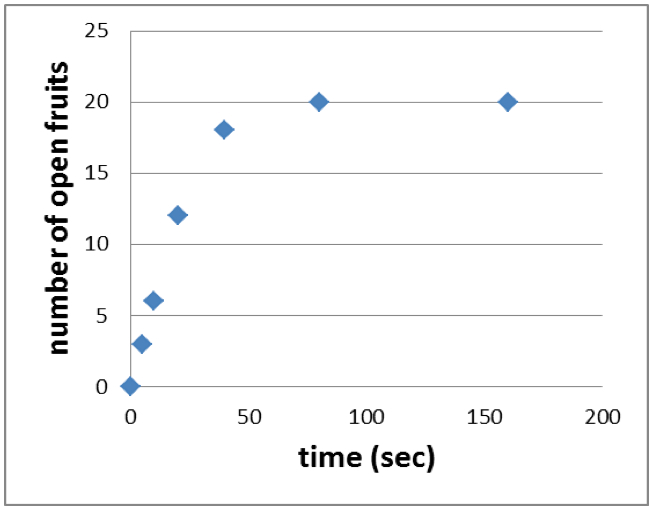
Figure 4. Cumulative agitation time is plotted against the number of open fruits
- Because during the following log transformation, you will run into trouble with the 0 data point, you add 1 (+1) to each time point (ending up with cumulative times of 6, 11, 21, 41, 81, 161).
- Then you apply the log10 to each time point to end up with a sigmoid graph like shown in Figure 5.
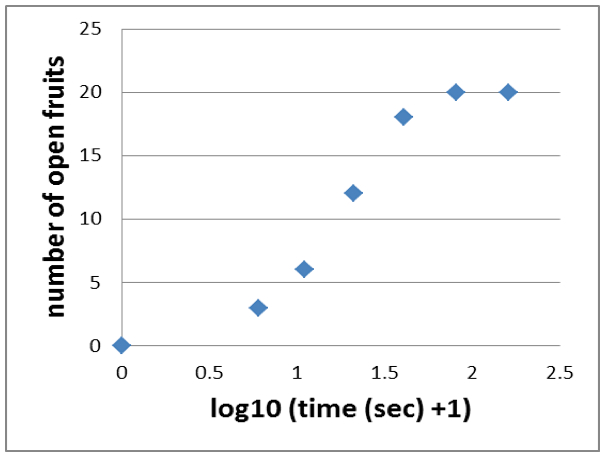
Figure 5. Applying the log10 to each time point results in a sigmoid graph
- Sigmoid graphs can be linearized with the logit function. Thus you apply the logit to the number of open fruits. Because the logit is not defined for 0 and 100% (in our case 0 and 20) you end up with a near linear relationship of only 4 data points (Figure 6).
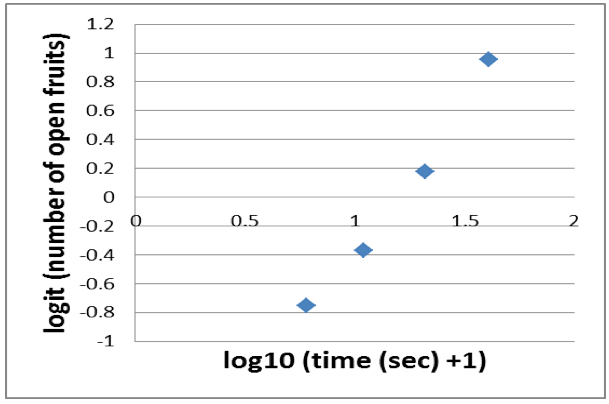
Figure 6. Near linear relationship derived from a logit transformation of the number of open fruits
- A linear slope is fitted to the data (for example using the Excel ‘Trendline’-function) which gives you a linear equation (Figure 7).
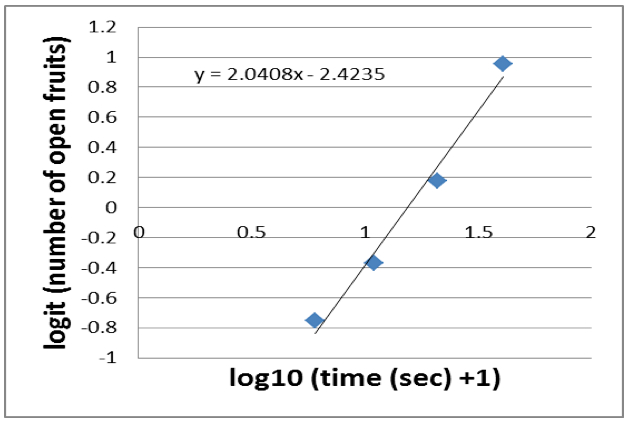
Figure 7. A linear slope is fitted to the data
- Now you want to calculate the halftime of dehiscence, which is the time point when half of the fruits (10 fruits) are open. The logit of 50% (in our case 10 fruits) equals 0. Thus, you have to calculate the x-intercept. In our example, the x-intercept is 2.4235/2.0408=1.1875.
- Finally, you just have to reverse the log transformation and the addition of 1 (see steps 10-11). Thus you calculate 101.1875 -1 = 14.3992, which is the dehiscence half-life in seconds.
- Repeat this measurement at least twice for each plant and calculate the mean half-life and standard deviation.
Acknowledgments
We thank Andreas Mühlhausen and Klaus Mummenhoff (Department of Botany, University of Osnabrück, Germany) for their kind cooperation in our project on fruit dehiscence and Thorsten Lenser for his help with data analysis. This protocol has been adapted based on previously published work (Morgan et al., 1998; Bruce et al., 2002; Arnaud et al., 2010). Our work was supported by a grant from the Deutsche Forschungsgemeinschaft to G.T. (TH 417/6-1).
References
- Arnaud, N., Girin, T., Sorefan, K., Fuentes, S., Wood, T. A., Lawrenson, T., Sablowski, R. and Ostergaard, L. (2010). Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev 24(19): 2127-2132.
- Bruce, D., Farrent, J., Morgan, C. and Child, R. (2002). PA-precision agriculture: determining the oilseed rape pod strength needed to reduce seed loss due to pod shatter. Biosyst Eng 81(2): 179-184.
- Lenser, T. and Theißen, G. (2013). Conservation of fruit dehiscence pathways between Lepidium campestre and Arabidopsis thaliana sheds light on the regulation of INDEHISCENT. Plant J 76(4): 545-556.
- Morgan, C., Bruce, D., Child, R., Ladbrooke, Z. and Arthur, A. (1998). Genetic variation for pod shatter resistance among lines of oilseed rape developed from synthetic B. napus. Field Crops Research 58(2): 153-165.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lenser, T. and Theißen, G. (2014). Quantifying Fruit Dehiscence Using the Random Impact Test (RIT). Bio-protocol 4(15): e1200. DOI: 10.21769/BioProtoc.1200.
Category
Plant Science > Plant developmental biology > Morphogenesis
Plant Science > Plant physiology > Abiotic stress
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










