- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Infectious Virus Yield Assay for Hepatitis E Virus
Published: Vol 4, Iss 15, Aug 5, 2014 DOI: 10.21769/BioProtoc.1195 Views: 11477
Reviewed by: Kanika GeraAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A SYBR Green-based Real Time RT-PCR Assay for Detection of the Emerging H7N9 Virus
Zheng Zhu and Lunbiao Cui
Jun 20, 2014 10104 Views

Quantification of HIV RNA and Human Herpesvirus DNA in Seminal Plasma
Milenka V. Vargas-Meneses [...] Sara Gianella
May 5, 2015 11130 Views

Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 3038 Views
Abstract
Hepatitis E virus (HEV) is one of the main causes of acute hepatitis worldwide. Infections are particularly severe in pregnant women and chronic hepatitis E is known to occur in immunocompromised patients. Current therapy (ribavirin or pegylated alpha interferon) has severe side effects and cannot be employed in all patients. In order to evaluate potential new inhibitors of HEV replication, a virus yield assay can be employed in which the amount of viral RNA progeny released into the culture medium is quantified by reverse-transcription quantitative PCR (RT-qPCR) (Debing et al., 2014).
Materials and Reagents
- HepG2/C3A human hepatoma cell line (ATCC, catalog number: CRL-10741 )
- Huh7 human hepatoma cell line (Japanese Collection of Research Bioresources, catalog number: JCRB0403 )
- HEV Kernow-C1 p6 plasmid (genotype 3 full-length genome; a kind gift from Suzanne Emerson, NIH) (Shukla et al., 2012)
- MluI restriction endonuclease with accompanying 10x buffer D (Promega Corporation, catalog number: R6381 )
- QIAquick gel extraction kit (QIAGEN, catalog number: 28704 )
- T7 RiboMAX large scale RNA production system (Promega Corporation, catalog number: P1300 )
- RNeasy mini kit (QIAGEN, catalog number: 74104 )
- ScriptCap m7G capping system (CELLSCRIPTTM, catalog number: C-SCCE0610 )
- Anti-fungal agent (2.5 µg/ml, Fungizone) (Life Technologies, catalog number: 15290-018 )
- Dulbecco’s modified Eagle’s medium with high glucose (DMEM) (Life Technologies, catalog number: 41965-039 )
- Fetal bovine serum (FBS) (not heat-inactivated) (Life Technologies, catalog number: 10270-106 )
- Opti-MEM I reduced serum medium (Life Technologies, catalog number: 31985-062 )
- Lipofectin transfection reagent (Life Technologies, catalog number: 18292-011 )
- Dulbecco’s phosphate-buffered saline (PBS) without Ca2+ and Mg2+ (Life Technologies, catalog number: 14190-094 )
- Penicillin/Streptomycin (PS) (10,000 IU/ml) (Life Technologies, catalog number: 15140-148 )
- CellTiter 96 AQueous MTS reagent powder [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] (Promega corporation, catalog number: G1111 )
- Phenazine methosulphate (PMS) (Sigma-Aldrich, catalog number: P9625 )
- Minimum essential medium (MEM) (no glutamine, no phenol red) (Life Technologies, catalog number: 51200-046 )
- NucleoSpin RNA virus kit (MACHEREY-NAGEL, catalog number: 740956 )
- One step qRT-PCR MasterMix Low Rox for probe assays (Kaneka Corporation, Eurogentec, catalog number: RT-QPRT-032XLR )
- Forward primer (5’-GGTGGTTTCTGGGGTGAC-3’) and reverse primer (5’- AGGGGTTGGTTGGATGAA-3’) (custom order) (Integrated DNA Technologies) (dissolved to a final concentration of 10 µM in buffer TE)
- Fluorescent probe (5’-FAM-TGATTCTCAGCCCTTCGC-MGBNFQ-3’ with FAM, 6-carboxyfluorescein; MGBNFQ, minor groove binder non-fluorescent quencher) (Life Technologies, custom order) (dissolved to a final concentration of 10 µM in buffer TE)
- qPCR DNA standard
The cloned cDNA target sequence is ligated into a plasmid (e.g. with the CloneJet PCR cloning kit, Thermo Fisher Scientific, catalog number: K1231 ) and miniprepped.
Note: The concentration is determined by spectrophotometry and a logarithmic dilution series is made in buffer TE over 6 orders of magnitude. Alternatively, the original Kernow-C1 p6 plasmid can be used to make such a dilution series.
- Tris (hydroxymethyl) aminomethane powder (Trizma base) (Sigma-Aldrich, catalog number: T1503 ) (dissolved to 1 M and pH 8.0 in H2O)
- Ethylenediaminetetraacetic acid disodium salt solution (EDTA, 0.5 M) (Sigma-Aldrich, catalog number: E7889 )
- MTS/PMS solution (see Recipes)
- Buffer TE (see Recipes)
Equipment
- Spectrophotometer to determine RNA concentrations (e.g. Thermo Fischer Scientific, NanoDrop, model: ND-1000 )
- Falcon transparent 6-well plate (Corning Incorporated, catalog number: 353046 )
- Falcon transparent 96-well plate (Corning Incorporated, catalog number: 353072 )
- 37 °C and 35 °C 5% CO2 cell culture incubators
- Centrifuge with temperature control
- Saphire² microplate reader for absorbance (Tecan Trading AG)
- PCR plate 96-well (SARSTEDT AG, catalog number: 72.1981.202 )
- MicroAmp optical adhesive film (Life Technologies, catalog number: 4311971 )
- ABI 7500 fast real-time PCR system (Life Technologies, catalog number: 4351107 )
Procedure
- Production of an infectious HEV stock from plasmid
- Five µg of the Kernow-C1 p6 plasmid is linearized in a 100 µl reaction containing 10 µl of 10x buffer D and 2.5 µl of MluI (10 units/µl) which is incubated at 37 °C for 2 h.
- The digested DNA is purified using the Qiaquick gel extraction kit according to the manufacturer’s instructions with final elution in 30 µl of buffer EB.
Note: To increase the yield, add the recommended amount of isopropanol to the mixture of the reaction and buffer QG. Other methods of DNA purification should be suitable as well.
- An in vitro transcription reaction is prepared with the T7 RiboMAX large scale RNA production system:
- 10 µl of 5x T7 transcription buffer
- 15 µl of NTPs (ATP, GTP, CTP, UTP; each at 25 mM)
- 20 µl of DNA template
- 5 µl of enzyme mix
Reaction is incubated at 37 °C for 5 h. Afterwards, 5 µl of the supplied RQ1 RNase-free DNase solution is added and incubated at 37 °C for 15 min.
- The produced RNA is purified with the RNeasy mini kit according to the manufacturer’s instructions with elution in 60 µl of RNase-free H2O (in 2 elutions of 30 µl each). RNA concentration is determined by spectroscopy.
- Sixty µg of RNA is capped with the ScriptCap m7G capping system according to the manufacturer’s instructions.
- The produced RNA is purified with the RNeasy mini kit according to the manufacturer’s instructions with elution in 60 µl of RNase-free H2O (in 2 elutions of 30 µl each). RNA concentration is determined by spectroscopy.
- Huh7 cells are seeded into a transparent 6-well plate at 2 x 105 cells per well in 2 ml of DMEM supplemented with 10% FBS.
- Plate is incubated in a 5% CO2 incubator at 37 °C for 24 h.
- To transfect one well, 1 µg of capped viral RNA is added to 100 µl of Opti-MEM. In another tube, 10 µl of Lipofectin transfection reagent is added to 100 µl of Opti-MEM. Both tubes are incubated at room temperature for 30 min. Then, the contents of both tubes are combined and incubated for another 10 min at room temperature. An additional 800 µl of Opti-MEM is added and the resulting solution is mixed well.
- The culture medium is carefully removed from cells seeded earlier by aspiration with a pipette and each well is washed once with 2 ml of DMEM (i.e. DMEM is added to the well, incubated for a few seconds while gently rocking the plate and removed by aspiration).
- To each well, 1 ml of transfection mixture is added.
- Plate is incubated in a 5% CO2 incubator at 37 °C for 5 h.
- Transfection medium is removed from each well and cell layers are washed once with 2 ml of PBS (see step A10).
- To each well, 2.5 ml of DMEM supplemented with 10% FBS and 1% PS is added.
Note: Optionally, amphotericin B can be added to the culture medium as an anti-fungal agent.
- Plate is incubated in a 5% CO2 incubator at 35 °C.
- Every 2-3 days (3x per week), 1 ml of culture medium is removed from each well and replaced with 1 ml of fresh DMEM supplemented with 10% FBS and 1% PS.
Note: These partial changes of the medium are essential to maintain cell viability over the extended incubation period.
- Twenty days post transfection, the culture medium is collected and centrifuged for 10 min at 1,000 x g at 4 °C. The supernatant is aliquoted and stored at -80 °C.
Note: The supernatant can be used directly as a viral stock or can be passaged once on HepG2/C3A cells for upscaling (similar to the protocol below). If required, the number of HEV RNA copies can be quantified by RT-qPCR as described below. In this case, a short RNase treatment is recommended prior to RNA extraction to remove remaining input RNA (the progeny viral RNA will be protected by the viral capsid).
- Five µg of the Kernow-C1 p6 plasmid is linearized in a 100 µl reaction containing 10 µl of 10x buffer D and 2.5 µl of MluI (10 units/µl) which is incubated at 37 °C for 2 h.
- Infectious virus yield assay
- HepG2/C3A cells are seeded into a transparent 6-well plate at 2 x 105 cells per well in 2 ml of DMEM supplemented with 10% FBS.
- Plate is incubated in a 5% CO2 incubator at 37 °C for 24 h.
- The HEV stock is thawed rapidly at 37 °C and diluted 1: 10 (or to the desired concentration) in DMEM supplemented with 10% FBS and 1% PS. The potential antiviral molecule is added to the inoculum, an equivalent amount of solvent (e.g. dimethylsulfoxide) is added to another well to serve as a cell control.
Note: Since HEV replication in vitro is rather poor and a high multiplicity of infection is required for a successful infection, the virus stock is not diluted very strongly. Addition of compound to the inoculum allows to evaluate effects on early events in the viral life cycle.
- Medium is removed from the HepG2/C3A cells and 1 ml of diluted HEV inoculum is added.
- Plate is incubated in a 5% CO2 incubator at 35 °C for 5 h.
- Inoculum is removed from each well and cell layers are washed 3 times with 2 ml of PBS.
- To each well, 2.5 ml of DMEM supplemented with 10% FBS and 1% PS is added. Compound is added to the desired concentration and an equivalent amount of solvent is added to the cell control well(s).
- Plate is incubated in a 5% CO2 incubator at 35 °C.
Note: Optionally, 150 µl of culture medium can be removed from each well after 1 h and stored at -80 °C for later RNA extraction and analysis. This sample allows to determine the baseline viral load.
- Every 2-3 days (3x per week), 1 ml of culture medium is removed from each well, transferred to a 1.5 ml tube and stored at -80 °C for later RNA extraction and analysis. The removed medium is replaced with 1 ml of fresh DMEM supplemented with 10% FBS and 1% PS and compound or control solvent. Plate is incubated further in a 5% CO2 incubator at 35 °C.
- Twenty days post transfection, the last medium samples are transferred to 1.5 ml tubes and the rest of the culture medium is removed.
- Cell layers are washed with 2 ml of PBS and 1 ml of a 1: 20 dilution of MTS/PMS solution in colorless MEM is added to each well.
- Plate is incubated in a 5% CO2 incubator at 37 °C for 2 h.
- One hundred µl of culture medium from each well is transferred to a transparent 96-well plate and absorbance read-out is performed in the microplate absorbance reader by determining the optical density (OD) at 498 nm.
- The remaining diluted MTS/PMS solution is removed from each well and cell layers are washed once with 2 ml of PBS.
- To each well, 350 µl of buffer RLT (included in the RNeasy mini kit) is added for cell lysis.
- Cell lysates are resuspended in buffer RLT, transferred to a 1.5 ml tube and vortexed vigorously for 1 min.
- RNA is extracted from cell lysates with the RNeasy mini kit according to the manufacturer’s instructions with final elution in 30 µl of RNase-free H2O.
Note: Alternatively, cell lysates can be stored at -80 °C after vortexing for extraction at a later time point.
- Total RNA concentration of extracted intracellular RNA is measured spectrophotometrically.
- To extract viral RNA from culture medium samples, samples are thawed at room temperature and viral RNA is extracted from 150 µl of culture medium with the Nucleospin RNA virus kit according to the manufacturer’s instructions.
Note: All culture medium samples can be extracted or only those of the preferred time points (e.g. the baseline and day 20 samples).
- For quantification of viral RNA, the following reaction mixture is made (for analysis of a single sample):
- 12.5 µl of One step qRT-PCR MasterMix
- 0.0625 µl of forward primer at 100 µM (final concentration: 250 nM)
- 0.0625 µl of reverse primer at 100 µM (final concentration: 250 nM)
- 0.25 µl of probe at 10 µM (final concentration: 100 nM)
- 7.0625 µl of RNase-free H2O
- 0.125 µl of Euroscript/RNase inhibitor
Note: Multiply the indicated amounts and volumes by the number of samples and cDNA samples to analyze and include an additional 10% extra.
- Twenty µl from this mixture is dispensed into each well of a PCR plate and 5 µl of RNA extract is added.
- To allow for absolute quantification, 5 µl of the dilution series of cloned cDNA targets is added to wells containing 20 µl of reaction mixture. In a last well, 5 µl of H2O is added to 20 µl reaction mixture (negative control).
- PCR plate is covered with an adhesive transparent cover. Plate is centrifuged shortly.
- RT-qPCR is performed with following temperature program:
- 30 min 48 °C
- 10 min 95 °C
- 15” 95 °C |
- 1 min 60 °C | x40
With the detector preset to FAM (520 nm) and data collection during the combined annealing-elongation step at 60 °C.
- HepG2/C3A cells are seeded into a transparent 6-well plate at 2 x 105 cells per well in 2 ml of DMEM supplemented with 10% FBS.
Representative data
- A representative example of the standard curve generated for the HEV RT-qPCR is shown below:
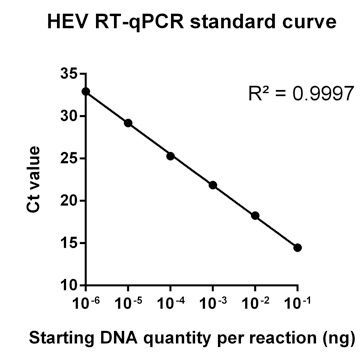
Figure 1. Representative example of the HEV RT-qPCR standard curve. This standard curve was generated using a series of 10-fold dilutions of the cloned target cDNA.
- An example of data generated by the method described here can be found below. Known HEV replication inhibitors ribavirin (RBV) and alpha interferon (IFNα) were tested (GE = genome equivalents, adapted from Debing et al., 2014).
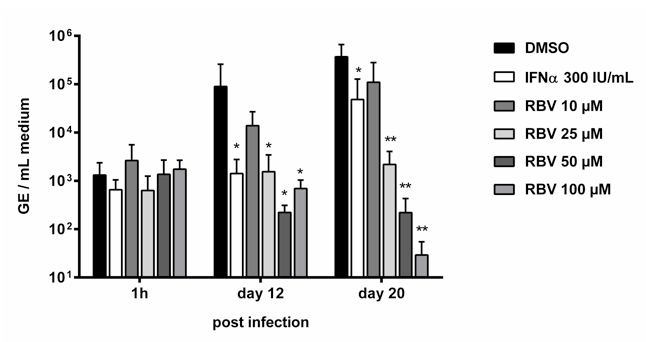
Figure 2. Antiviral activities of interferon alpha (IFNα) and different concentrations of ribavirin (RBV) were assessed in Huh7 cells using the infectious virus yield assay with RT-qPCR detection of viral RNA. *, P < 0.05; **, P < 0.01. GE, genome equivalents. Adapted from Debing et al. (2014).
Notes
- In general, Huh7 cells are more permissive to transfection than HepG2/C3A cells, while the latter are infected more efficiently and thus result in higher yields after infection. However, it is possible to transfect and infect both cell lines with the protocol outlined above, although differences in efficiency and yield may be observed.
- The quantities obtained by RT-qPCR are based on the concentrations of the standard dilution series and are thus expressed in ng/µl. Using the molecular weight of the plasmid standard and Avogadros number, the number of copies can be calculated. For intracellular RNA, it is advised to normalize the obtained copy number for the total cellular RNA (as determined spectrophotometrically) or for a house-keeping gene which mRNA levels are unaffected by HEV infection or compound treatment.
- To calculate cell viability, the following formula can be employed:
% Viability = ODcompound/ODCC x 100
Recipes
- MTS/PMS solution
- Two g of MTS powder is dissolved in 1 L of PBS and stirred for 15 min.
- Next, 46 mg of PMS powder is added (pH should be between 6-6.5.).
- Solution is filtered, aliquoted and stored at -20 °C.
- Two g of MTS powder is dissolved in 1 L of PBS and stirred for 15 min.
- Buffer TE
To 25 ml of nuclease-free H2O, add
250 µl of a 1 M Tris solution (pH 8.0) (final concentration: 10 mM)
50 µl of a 0.5 M EDTA solution (final concentration: 1 mM)
Acknowledgments
This protocol was adapted from Debing et al. (2014) and is partially based on earlier work by Shukla et al. (2012). Primer and probe sequences are derived from Jothikumar et al. (2006). Yannick Debing is a fellow of the Research Foundation-Flanders (FWO). This work was supported by KU Leuven Geconcerteerde Onderzoeksacties (GOA/10/014) and by EU FP7 project SILVER (260644).
References
- Debing, Y., Emerson, S. U., Wang, Y., Pan, Q., Balzarini, J., Dallmeier, K., Neyts, J. (2014). Ribavirin inhibits in vitro hepatitis E virus replication through depletion of cellular GTP pools and is moderately synergistic with alpha interferon. Antimicrob Agents Chemother 58(1): 267-273.
- Jothikumar, N., Cromeans, T.L., Robertson, B.H., Meng, X.J., Hill, V.R. (2006) A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J Virol Methods 131(1): 65-71.
- Shukla, P., Nguyen, H.T., Faulk, K., Mather, K., Torian, U., Engle, R.E., Emerson, S.U. (2012) Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination. J Virol 86(10): 5697-5707.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Debing, Y., Dallmeier, K. and Neyts, J. (2014). Infectious Virus Yield Assay for Hepatitis E Virus. Bio-protocol 4(15): e1195. DOI: 10.21769/BioProtoc.1195.
Category
Microbiology > Microbe-host interactions > Virus
Microbiology > Microbial genetics > RNA > qRT-PCR
Molecular Biology > RNA > RNA detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











