- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of Tomato Fruit Chromoplasts and Determination of ATP Levels
Published: Vol 4, Iss 15, Aug 5, 2014 DOI: 10.21769/BioProtoc.1192 Views: 11159
Reviewed by: Beery YaakovTie Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1474 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3209 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1824 Views
Abstract
It has recently been reported that tomato fruit chromoplasts can synthesize ATP de novo using an ATP synthase complex harboring an atypical γ-subunit which is also present in a variety of plant species. However many aspects related with the biochemical processes underlying this process remain largely unknown. Here we describe detailed protocols for the isolation of tomato fruit chromoplasts and the determination of ATP levels (end-point measurements) and ATP synthesis rates (kinetic measurements) in these organelles using bioluminescent luciferin/luciferase based assays.
Keywords: ChromoplastMaterials and Reagents
- Red tomato fruits (MicroTom or Ailsa Craig, harvested 5-7 and 7-9 days after breaker stage, respectively)
- Ultrapure water
- NaCl (Sigma-Aldrich, catalog number: S7653 )
- Trizma® base (Tris) (Sigma-Aldrich, catalog number: T6066 )
- HCl (Merck KGaA, catalog number: 100317 )
- Sorbitol (Sigma-Aldrich, catalog number: S1878 )
- MgCl2 (Sigma-Aldrich, catalog number: M2670 )
- KCl (Sigma-Aldrich, catalog number: P9541 )
- L-Ascorbic acid (Sigma-Aldrich, catalog number: 255564 )
- L-Cysteine (Sigma-Aldrich, catalog number: C7352 )
- DL-Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 43819 )
- Sucrose (Sigma-Aldrich, catalog number: S8501 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A7906 )
- MnCl2 (Sigma-Aldrich, catalog number: M3634 )
- KH2PO4 (Merck KGaA, catalog number: 104873 )
- β-Nicotinamide adenine dinucleotide 2’-phosphate reduced tetrasodium salt hydrate (NADPH) (Sigma-Aldrich, catalog number: N7505 )
- β-Nicotinamide adenine dinucleotide phosphate hydrate (NADP+) (Sigma-Aldrich, catalog number: N5755 )
- β-Nicotinamide adenine dinucleotide reduced disodium salt hydrate (NADH) (Sigma-Aldrich, catalog number: N8129 )
- β-Nicotinamide adenine dinucleotide hydrate (NAD+) (Sigma-Aldrich, catalog number: N1636 )
- Flavin adenine dinucleotide disodium salt hydrate (FAD) (Sigma-Aldrich, catalog number: F6625 )
- Sodium pyruvate (Sigma-Aldrich, catalog number: P2256 )
- ATP Bioluminescence Assay Kit HS II (Roche Diagnostics, catalog number: 11699709001 )
- ENLITEN® rLuciferase/Luciferin Reagent (Promega Corporation, catalog number: FF2021 )
- RC DC Protein Assay kit (Bio-Rad Laboratories, catalog number: 500-0122 )
- Adenosine 5’-diphosphate sodium salt (ADP) (Sigma-Aldrich, catalog number: A2754 )
- Adenosine 5’-triphosphate disodium salt hydrate (ATP) (Sigma-Aldrich, catalog number: A2383 )
- P1, P5-di(adenosine-5’) pentaphosphate pentasodium salt (DAPP) (Sigma-Aldrich, catalog number: D4022 )
- 2-[(2-Hydroxy-1,1-bis(hydroxymethyl)ethyl)amino]ethanesulfonic acid, N-[Tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid (TES) (Sigma-Aldrich, catalog number: T1375 )
- 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid, N-(2-Hydroxyethyl)piperazine-N-(2-ethanesulfonic acid) (HEPES) (Sigma-Aldrich, catalog number: H4034 )
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884 )
- Tween 20 (Sigma-Aldrich, catalog number: P1379 )
- Miracloth (pore size of 22-25 µm) (Calbiochem®, catalog number: 475855 )
- 1 mM DAPP (stored at -20 °C)
- 10 mM FAD (stored at -80 °C)
- 100 mM NADH (stored at -80 °C)
- 100 mM NAD+ (stored at -80 °C)
- 100 mM NADPH (stored at -80 °C)
- 100 mM NADP+ (stored at -80 °C)
- 10 µM ATP standard solutions (stored at -80 °C)
- 0.1 µM ATP standard solutions (stored at -80 °C)
- 0.001 µM ATP standard solutions (stored at -80 °C)
- 1 M DTT (stored at -20 °C)
- 0.5 mM ADP
- Washing solution (see Recipes)
- 0.5 M Tris-HCl buffer (see Recipes)
- 15% sucrose solution (see Recipes)
- 30% sucrose solution (see Recipes)
- 40% sucrose solution (see Recipes)
- 50% sucrose (see Recipes)
- Buffer A (see Recipes)
- Buffer B (see Recipes)
- Buffer C (see Recipes)
- Buffer D (see Recipes)
- Buffer E (see Recipes)
Equipment
- Glomax® 96 Microplate Luminometer w/Dual Injectors (Promega Corporation, catalog number: E6521 )
- Centrifuge Avanti J-E (Beckman Coulter)
- Ultracentrifuge Optima L-100xPI (Beckman Coulter)
- SW 28 swinging-bucket rotor (Beckman Coulter) or equivalent
- JA-14 rotor (Beckman Coulter) or equivalent
- JA-20 rotor (Beckman Coulter) or equivalent
- Polypropylene centrifuge tubes (250 ml) (Beckman Coulter)
- Polypropylene centrifuge tubes (30 ml) (Beckman Coulter)
- Polyallomer centrifuge tubes for SW 28 rotor (Beckman Coulter)
- Waring blender 8011S (Dynamics Corporation of America)
- Support stand, clamp holder and extension clamp
- Glass beakers of different volume capacity
- 96-well microplates white (Porvair Sciences, catalog number: 204003 )
- Bench orbital shaker
- Micropipettes
- Heat block
- Benchtop microcentrifuge
- Gauze (1 m x 1 m)
Software
- Microsoft Excel
Procedure
- Chromoplast isolation
- Prepare in advance the buffers for chromoplast isolation (buffers A and B) and the sucrose solutions for ultracentrifugation in discontinuous gradients.
- Wash about 300 g tomato fruits with washing solution for 15 min.
- Cut fruits and remove seeds and the gelatinous material of the locular cavities. Store the pericarp tissue in a glass beaker placed on ice.
- Cut the pericarp tissue in small pieces (approximately 1 cm2) with a razor blade.
- Weigh the pericarp tissue and place in the cold Waring blender jar.
- Add two volumes of ice-cold buffer A.
- Homogenize with three pulses of 1 sec each at low speed.
- Filter the homogenate through 8 layers of gauze and then through 2 layers of Miracloth. Transfer the flow through to two polypropylene centrifuge tubes of 250 ml. Keep samples on ice.
- Centrifuge at 200 x g for 2 min at 4 °C.
- Carefully transfer the supernatant to new polypropylene centrifuge tubes of 250 ml.
- Centrifuge at 5,000 x g for 10 min at 4 °C.
- Discard the supernatant and gently resuspend the pellets in 25 ml of ice-cold buffer B.
- Centrifuge at 5,000 x g for 10 min at 4 °C.
- Gently resuspend the chromoplast pellet in 4 ml of ice-cold buffer B using a 1 ml micropipette (cut the tip to increase its diameter for not harming the chromoplasts).
- In advance, prepare the sucrose differential density blocks in two 35 ml SW28 ultracentrifuge tubes by carefully loading the corresponding sucrose solutions in the following order (from bottom to top): 8 ml of 50% sucrose solution, 8 ml of 40% sucrose solution, 8 ml of 30% sucrose solution and 8 ml of 15% sucrose solution. Keep at 4 °C.
- Carefully load 2 ml of the resuspended chromoplast sample on top of the 15 % sucrose solution.
- Centrifuge at 50,000 x g for 60 min at 4 °C in a SW28 rotor (mild brake).
- Carefully remove the tubes from the buckets and hold them using an extension clamp.
- Carefully harvest the chromoplasts present in the 30-40% interface using a Pasteur pipette and transfer into a prechilled 30 ml centrifuge tube.
- Add one volume of ice-cold buffer B and centrifuge at 5,000 x g for 10 min at 4 °C.
- Resuspend chromoplasts in the appropriate buffer according to the protocols used for ATP measurements described in steps B and C.
- Prepare in advance the buffers for chromoplast isolation (buffers A and B) and the sucrose solutions for ultracentrifugation in discontinuous gradients.
- Measurement of ATP levels (end point measurement)
- Resuspend chromoplasts (step A21) in 750 µl of ice-cold buffer C by gentle pipetting. Samples should be kept continuously on ice.
- Aliquot the resuspended chromoplasts into microcentrifuge tubes (30 µl per tube) and incubate samples at room temperature (23 °C) on a bench orbital shaker for as long as needed according to the experiment (usually 10 to 60 min). This step allows chromoplasts to perform the biochemical reactions needed for the de novo synthesis of ATP. At the end of the incubation, samples can be processed immediately or stored at -80 °C.
- Add 90 µl of the “Cell Lysis Reagent” (contained in the kit “ATP Bioluminescence Assay Kit HS II”) to each tube, mix and incubate the samples for 2 min at 95 °C in a heat block, then transfer the samples on ice.
- Centrifuge the samples for 3 min in a microcentrifuge at high speed and room temperature to pellet chromoplasts debris. Transfer the supernatants to fresh microcentrifuge tubes. Keep samples on ice.
- Transfer 50 µl from each sample to a 96-well microplate. Keep samples on ice.
- Perform the ATP determination reactions using the Glomax 96 Microplate Luminometer. For these experiments only one injector is used, primed with the “Luciferase reagent” included in the kit “ATP Bioluminescence Assay Kit HS II” and prepared in advance following the instructions provided by the supplier. The instrument is set to add automatically 50 µl of the “Luciferase reagent” to each sample and to start the measurements after 1 sec delay. The final outcome values of the instrument show relative luminescence (light) of the samples, which expresses ATP content according to the reaction: D-luciferin + ATP + O2 → oxyluciferin + PPi + AMP + CO2 + light. The values of the blank samples (samples without chromoplasts, but only the buffer C) have to be subtracted from the sample values.
Note: The above described method using the “ATP Bioluminescence Assay Kit HS II” kit allows the determination of relative ATP values and the comparison of the ATP content of different samples according to the luminescence observed values. Furthermore, absolute ATP values can be measured using the same kit and method. For this purpose, the ATP standard (contained in the kit) has to be prepared, according to the kit manual, in serial dilutions of known ATP concentration. Next, the luminescence of these serial dilutions has to be determined using the Glomax 96 Microplate Luminometer to generate an ATP standard curve. Comparison of the luminescence values of the samples with those of the ATP standard curve can help for the estimation of absolute ATP values of the samples of interest. The estimated ATP amount can be referred to chromoplast protein content (see step D).
- Resuspend chromoplasts (step A21) in 750 µl of ice-cold buffer C by gentle pipetting. Samples should be kept continuously on ice.
- Measurement of ATP synthesis rates (kinetics measurements)
- Reconstitute the “ENLITEN® rLuciferase/Luciferin Reagent” following the instructions provided by the supplier. The reagent has to be stored at -20 °C in 2 ml aliquots for no more than two weeks.
- Resuspend chromoplasts (step A21) in 500 µl of buffer D by gentle pipetting. Keep chromoplast samples on ice and start the ATP measurements within the first 15 min after chromoplast isolation.
- Prime injector 1 of the luminometer with 0.5 mM ADP. Then select the “kinetic protocol” with injector 1 and adjust the settings as indicated: i) injector volume: 40 µl, ii) delay after injection: 0.4 sec, and iii) integration time: 60 sec.
- Allow the following reagents to equilibrate at room temperature for approximately 5 min. Then, add the reagents in this order into the wells of a 96-well microplate:Note: DAPP is an inhibitor of adenylate kinase that does not affect the ATP synthase.
rLuciferase/Luciferin Reagent 80 µl Buffer E 80 µl DAPP 4 µl NADH 100 mM and NAD+ 100 mM
(or NADPH 100 mM and NADP+ 100 mM)2 µl of each Chromoplasts resuspended in buffer D 20 µl - Place the microplate in the luminometer and start the “kinetic protocol”. The injector should inject automatically 40 µl of 0.5 mM ADP and light emission data should be collected during 60 sec for each well.
- To prepare the ATP calibration curve, select the “non-injection kinetic protocol” in the luminometer and adjust the following settings: i) delay in the first measurement: 0 sec, and ii) integration time: 30 sec.
- Add the following components in the appropriate wells of the microplate (at room temperature):Note: Chromoplasts are added because they quench part of the light emitted in the luciferase reaction.
rLuciferase/luciferin Reagent 80 µl Buffer 80 µl Ultrapure water 20 µl DAPP 4 µl Corresponding ATP standard (10 µM, 0.1 µM or 0.001 µM ATP) 20 µl Chromoplasts resuspended in buffer D 20 µl - Place the plate in the luminometer and start the “non-injection kinetic protocol”.
- Data analysis: Relative luminescence data will appear in an Excel spreadsheet. Calculate the ATP standard curve equation using the Excel functions and apply this equation to obtain the ATP concentration of all samples at each measurement time (sec). Then, the ATP synthesis rate can be represented in a graph (Y-axis: nmol ATP; X-axis: sec) and can be expressed as nmol ATP x mg protein-1 x sec-1 performing the appropriate calculations.
Note: Data from the first 5 sec of kinetics measurements are discarded because they are usually erratic.
- Reconstitute the “ENLITEN® rLuciferase/Luciferin Reagent” following the instructions provided by the supplier. The reagent has to be stored at -20 °C in 2 ml aliquots for no more than two weeks.
- Determination of protein content of chromoplast samples
- Aliquots of 50 µl of chromoplast samples resuspended in buffer C or D are supplemented with Tween-20 at 1% final concentration and incubated at 4 °C in an orbital shaker for 30 min.
- Process the samples using the RC DC Protein Assay kit following the indications provided by the supplier.
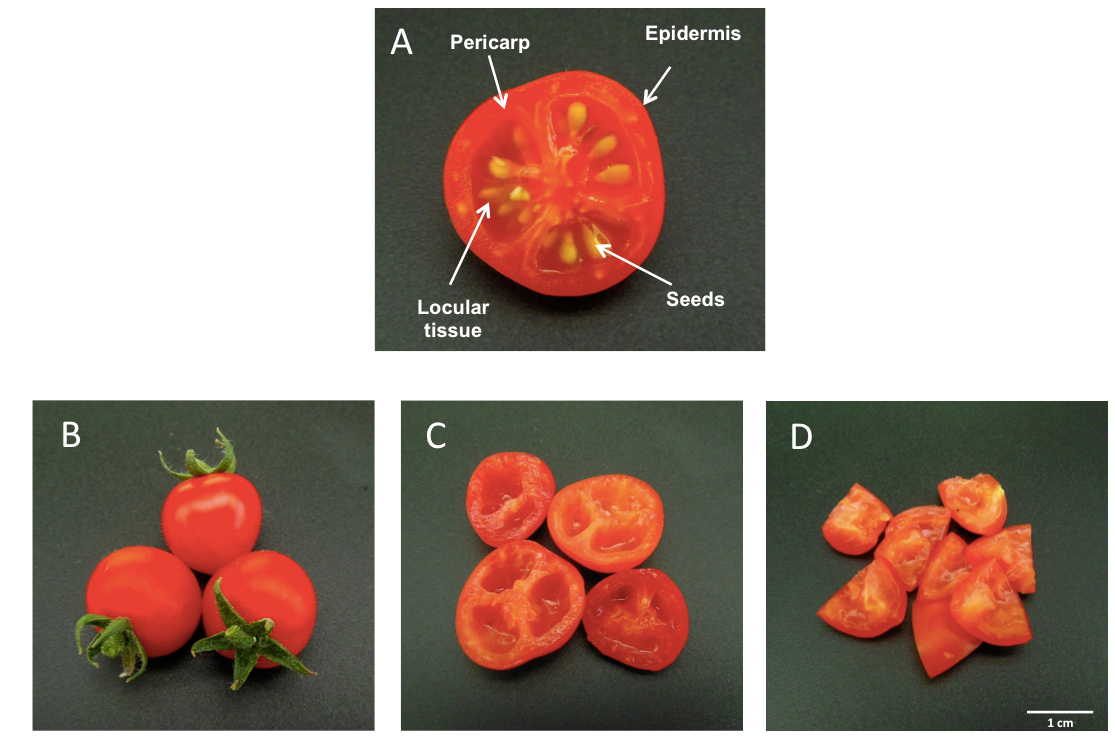
Figure 1. Transversal section of a tomato fruit (var. MicroTom). (A). Preparation of fruit samples prior to homogenization (B, C and D).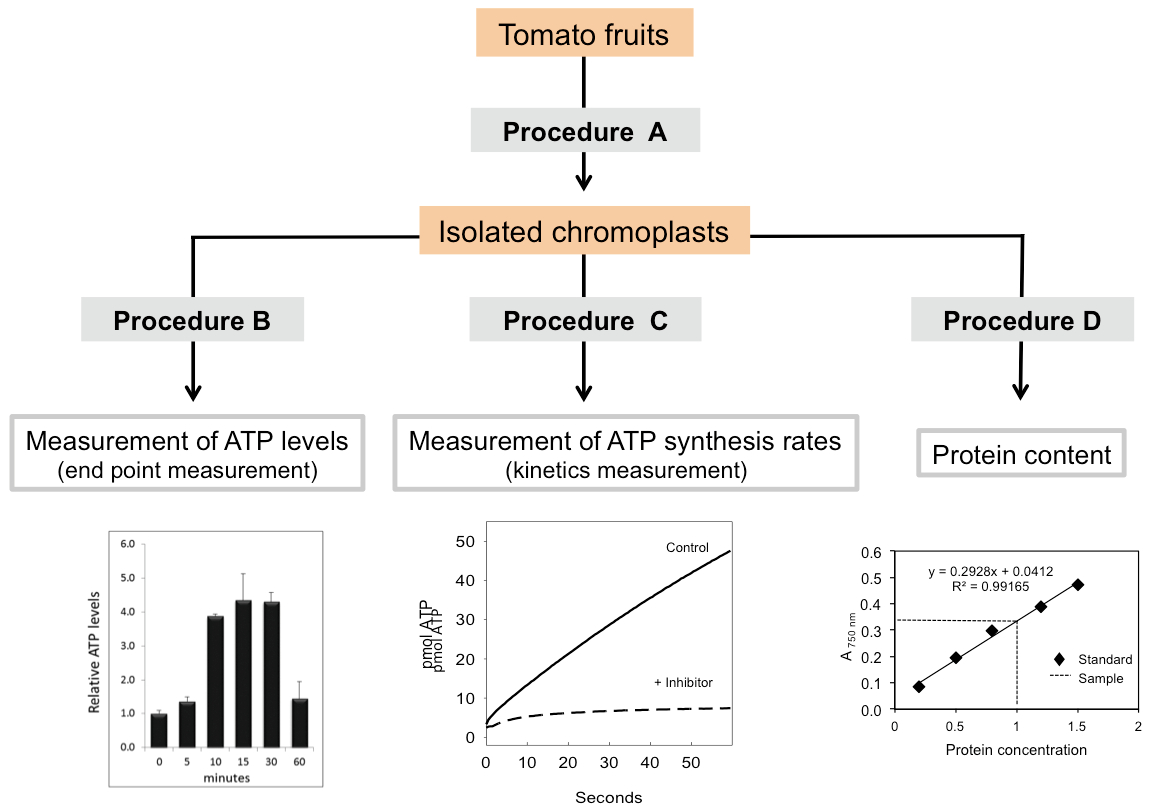
Figure 2. Schematic overview of the procedures included in this protocol showing the typical results obtained in each case
- Aliquots of 50 µl of chromoplast samples resuspended in buffer C or D are supplemented with Tween-20 at 1% final concentration and incubated at 4 °C in an orbital shaker for 30 min.
Recipes
- Washing solution
Mix 12.5 g of NaCl in 500 ml of distilled water - 0.5 M Tris-HCl (pH 7.4) buffer
Stored at 4 °C - 15% sucrose solution (50 ml) (prepare before use)
Mix 7.5 g of sucrose with 5 ml 0.5 M Tris (pH 7.4) buffer and 50 µl of 1 M DTT
Complete to 50 ml with ultrapure water - 30% sucrose solution (50 ml) (prepare before use)
Mix 15 g of sucrose with 5 ml 0.5 M Tris (pH 7.4) buffer and 50 µl of 1 M DTT
Complete to 50 ml with ultrapure water - 40% sucrose solution (50 ml) (prepare before use)
Mix 20 g of sucrose with with 5 ml 0.5 M Tris (pH 7.4) buffer and 50 µl of 1 M DTT
Complete to 50 ml with ultrapure water - 50% sucrose (50 ml) (prepare before use)
Mix 25 g of sucrose, 5 ml 0.5 M Tris (pH 7.4) buffer and 50 µl of 1 M DTT
Complete to 50 ml with ultrapure water - Buffer A (prepare fresh before use)
100 mM Tris-HCl (pH 8.2)
330 mM sorbitol
2 mM MgCl2
10 mM KCl
8 mM EDTA
10 mM L-ascorbic acid
5 mM L-cysteine
0.2% BSA (can be stored at 4 °C)
Just before use add DTT (to 1 mM final concentration)
Finally, add PVPP to 1% and mix. - Buffer B (prepare fresh before use)
Buffer A without PVPP - Buffer C (prepare fresh prior to use)
100 mM HEPES-HCl (pH 7.4)
10 mM MgCl2
2 mM MnCl2
10 mM KH2PO4
1 mM NADPH
1 mM NADP+
20 μM FAD
2 mM pyruvate
330 mM sorbitol - Buffer D
100 mM HEPES-HCl (pH 7.4)
10 mM MgCl2
2 mM MnCl2
10 mM KH2PO4
330 mM sorbitol
Before use add FAD to 10 µM final concentration
Stored at -20 °C - Buffer E
10 mM TES-HCl (pH 7.4)
600 mM sorbitol
2 mM MgCl2
25 mM KH2PO4
0.33 mM EDTA
Stored at -20 °C
Note: All buffers and solutions are prepared with ultrapure water.
Acknowledgments
This protocol has been adapted from methods previously described in Angaman et al. (2012) and Pateraki et al. (2013). This work has been supported by grants from the Spanish Ministerio de Ciencia e Innovación (BIO2009-09523) and Ministerio de Economía y Competitividad (AGL2013-43522-R), both including FEDER Funds, the Spanish Consolider-Ingenio 2010 Program (CSD2007-00036 Centre for Research in Agrigenomics) and the Generalitat de Catalunya (2009SGR0026). Marta Renato is a recipient of a predoctoral fellowship from the Spanish Ministerio de Educación, Cultura y Deporte.
References
- Angaman, D. M., Petrizzo, R., Hernandez-Gras, F., Romero-Segura, C., Pateraki, I., Busquets, M. and Boronat, A. (2012). Precursor uptake assays and metabolic analyses in isolated tomato fruit chromoplasts. Plant Methods 8(1): 1.
- Pateraki, I., Renato, M., Azcón‐Bieto, J. and Boronat, A. (2013). An ATP synthase harboring an atypical γ–subunit is involved in ATP synthesis in tomato fruit chromoplasts. Plant J 74(1): 74-85.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hernández-Gras, F., Petrizzo, R., Pateraki, I., Renato, M., Angaman, M., Azcón-Bieto, J. and Boronat, A. (2014). Isolation of Tomato Fruit Chromoplasts and Determination of ATP Levels. Bio-protocol 4(15): e1192. DOI: 10.21769/BioProtoc.1192.
Category
Plant Science > Plant cell biology > Organelle isolation
Plant Science > Plant biochemistry > Protein > Activity
Cell Biology > Organelle isolation > Fractionation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










