- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Dye Release Experiments with Dextran Loaded Vesicles
Published: Vol 4, Iss 14, Jul 20, 2014 DOI: 10.21769/BioProtoc.1190 Views: 10468
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Fluorescence Dequenching-based Liposome Leakage Assay to Measure Membrane Permeabilization by Pore-forming Proteins
Javier Aguilera [...] Jianjun Sun
May 20, 2021 7126 Views

Flotation Assay With Fluorescence Readout to Study Membrane Association of the Enteroviral Peripheral Membrane Protein 2C
Kasturika Shankar [...] Lars-Anders Carlson
Apr 5, 2025 1623 Views

Lipid-Mediated Sequential Recruitment of Proteins Via Dual SLIPT and Dual SLIPTNVOC in Live Cells
Kristina V. Bayer and Richard Wombacher
Nov 5, 2025 1604 Views
Abstract
Dye release experiments are a widely used method to assess the interactions between membrane-active molecules and lipid membranes. Of particular interest is the ability to assess the degree of the lipid bilayer perturbation by simultaneously encapsulating dye of different sizes, such as dextrans grafted with a chromophore. In this assay, dextran linked to rhodamine or fluorescein are both encapsulated in lipid vesicles to allow quantifying the leakage of each dextran individually from a single sample. For instance, the size evaluation of the lipid pore formed by an antimicrobial peptide has been recently achieved using this protocol (Sani et al., 2013).
Keywords: Model membranesMaterials and Reagents
- Rhodamine-dextran (RD) of different molecular weights [for instance 40 kDa RD molecular weight (RD-40)]
- Fluorescein-dextran (FD) of different molecular weights [for instance 4.4 kDa FD (FD-4)]
- Tris.HCl
- NaCl
- MQ-water
- Triton-X100
- Peptide of interest e.g. Maculatin 1.1 (Sani et al., 2013)
- Lipids of interest e.g. 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG), 1',3'-bis[1,2-dioleoyl-sn-glycero-3-phospho]-sn-glycerol (TOCL) (Sani et al., 2013)
Equipment
- Polycarbonate membrane filters (200 nm diameter)
- Extruder (for instance Avanti Mini-extruder, Alabaster)
- Bench centrifuge
- Spectrofluorimeter and quartz cuvette
Procedure
- Stock preparation
- Prepare a Tris.HCl + X NaCl buffer (buffer A) and a Tris.HCl + (X-Dextran concentration) NaCl buffer (buffer B) at desired pH, with X being the NaCl concentration.
- Prepare a peptide stock solution (The concentration will depend on the peptide solubility and the required lipid to peptide molar ratio under investigation.) in buffer A. Glass vials are recommended.
- Prepare a stock solution of equimolar RD and FD in buffer B.
- Co-solubilize lipids in chloroform/methanol (3: 1 v/v), remove the organic solvents under vacuum using a rotary evaporator. Rehydrate the lipids in Milli-Q water and lyophilize.
- Resuspend the require mass of fluffy lipid powder with the dextran stock solution. A 15 mM lipid stock solution usually allows practical dilution to reach required lipid to peptide molar ratio, but will be dependent on the peptide stock concentration. Perform 3-5 freeze-thaw cycles to homogenise the liposome dispersion, usually without a time delay between cycles, by dipping the solution in liquid nitrogen and then melting above the lipid fluid phase transition for ~ 5 min.
- Extrude 10 times through an Extruder using 0.2 mm pore size polycarbonate filters to produce large unilamellar vesicles (LUV) of 200 nm diameter. Extrusion must be performed above the gel-to-fluid lipid phase transition to avoid vesicle aggregation or demixing of heterogeneous lipid compositions, if used.
- Remove un-encapsulated dye from LUV dispersions by centrifuging three times at 22,000 x g for 30 min at 25 °C. After each centrifugation, remove the supernatant and replace with an equal volume of fresh buffer B solution and mixed gently.
- Determine the lipid concentration of washed LUV dispersions in triplicate using the phosphorus assay of Sani et al. (2013).
- Prepare a Tris.HCl + X NaCl buffer (buffer A) and a Tris.HCl + (X-Dextran concentration) NaCl buffer (buffer B) at desired pH, with X being the NaCl concentration.
- Sample preparation
- Equilibrate the LUV dispersion and peptide solution at the desired temperature.
- Depending on fluorimeter performance, prepare an adequate dextran-encapsulated solution, ~ 200 μM gives a high signal in our experience.
- Add the require amount of peptide to obtain the lipid to peptide molar ratio (L/P) of interest. For reproducibility and statistical analysis, it is best to prepare the sample in triplicate.
- Produce negative control by adding buffer B instead of the peptide solution. A non-membrane active peptide or protein, such as bovine serum albumim, can also be used.
- Produce positive control by adding 0.5% Triton X-100 (Tx).
- Incubate the samples for 30 min at the desired temperature.
- Centrifuge the samples at 22,000 x g for 30 min.
- Recover the supernatant, with care to leave the pellet unperturbed. Usually, 500 μl sample are prepared and 300 μl recovered for analysis.
- Transfer the supernatant to a quartz cuvette.
- Equilibrate the LUV dispersion and peptide solution at the desired temperature.
- Fluorescence measurement
- Record the fluorescence emission of RD (then FD) using an excitation wavelength set at 550 nm (then 480 nm). Fluorescence emission is recorded from 560 to 650 nm using adequate integration time and PMT voltage. Triplicate the measurements.
- The amount of fluorescence is determined by integrating the area under the emission curve. The % release of each sample is calculated by using the following equation:
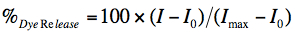 where, I is the fluorescence intensity of peptide treated vesicle supernatant, I0 obtained from the peptide-free sample (negative control) supernatant, and Imax from the Tx treated supernatant (positive control).
where, I is the fluorescence intensity of peptide treated vesicle supernatant, I0 obtained from the peptide-free sample (negative control) supernatant, and Imax from the Tx treated supernatant (positive control).
- Record the fluorescence emission of RD (then FD) using an excitation wavelength set at 550 nm (then 480 nm). Fluorescence emission is recorded from 560 to 650 nm using adequate integration time and PMT voltage. Triplicate the measurements.
Acknowledgments
None.
References
- Anderson, R. L. and Davis, S. (1982). An organic phosphorus assay which avoids the use of hazardous perchloric acid. Clinica Chimica Acta 121(1): 111-116.
- Sani, M. A., Whitwell, T. C., Gehman, J. D., Robins-Browne, R. M., Pantarat, N., Attard, T. J., Reynolds, E. C., O'Brien-Simpson, N. M. and Separovic, F. (2013). Maculatin 1.1 disrupts Staphylococcus aureus lipid membranes via a pore mechanism. Antimicrob Agents Chemother 57(8): 3593-3600.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sani, M., O’Brien-Simpson, N. M. and Separovic, F. (2014). Dye Release Experiments with Dextran Loaded Vesicles. Bio-protocol 4(14): e1190. DOI: 10.21769/BioProtoc.1190.
Category
Microbiology > Microbial biochemistry > Lipid
Biochemistry > Lipid > Lipid binding
Biochemistry > Protein > Interaction > Protein-lipid interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










