- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Chromatin Fractionation Assay in Fission Yeast
Published: Vol 4, Iss 14, Jul 20, 2014 DOI: 10.21769/BioProtoc.1185 Views: 12556
Reviewed by: Kanika GeraBelen SanzAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

MNase Digestion for Nucleosome Mapping in Neurospora
Cigdem Sancar [...] Michael Brunner
Jun 5, 2016 10314 Views

Using CRISPR/Cas9 for Large Fragment Deletions in Saccharomyces cerevisiae
Huanhuan Hao [...] Liping Zhang
Jul 20, 2017 14252 Views

Method for Multiplexing CRISPR/Cas9 in Saccharomyces cerevisiae Using Artificial Target DNA Sequences
Rachael M. Giersch and Gregory C. Finnigan
Sep 20, 2017 13376 Views
Abstract
The protein recruitment onto chromatin is a critical process for DNA metabolism, including DNA replication, DNA repair and DNA recombination. Especially DNA modification enzymes and checkpoint proteins are loaded onto DNA damage sites in a context-dependent manner. In our recent study (Kunoh and Habu, 2014), the chromatin association of Pcf1, a large subunit of Chromatin Assembly Factor-1 (CAF-1), was monitored after exposure of cells to hydroxyurea which slowed down the DNA replication. Results of the chromatin fractionation assay provided evidence that Pcf1 was recruited to chromatin upon DNA replication stress. A similar procedure enabled to reveal the chromatin association of Orp1, Mcm proteins, and Swi6 (Sadaie et al., 2008; Ogawa et al., 1999). This assay allows us to fractionate chromatin-bound and -unbound proteins from living cells. The following immunoblot of the respective fractions provides the information concerning the chromatin binding status of our target proteins.
Keywords: ChromatinMaterials and Reagents
- Yeast strain (Schizosaccharomyces pombe)
- Flask (IWAKI PUMPS, catalog number: 4980FK500 )
- Conical tube (BD Biosciences, Falcon®, catalog number: 2070 )
- 1.5 ml microcentrifuge tube (Eppendorf, catalog number: 022364111 )
- Lysing enzymes from Trichoderma harzianum (Sigma-Aldrih, catalog number: L1412 )
- Zymolyase 100T (Seikagaku Corporation, catalog number: 120493-1 )
- Complete Mini (Roche Diagnostics, catalog number: 11836153001 )
- Anti-GFP antibody (Roche Diagnostics, catalog number: 11814460001 )
- Anti-histone H3 antibody (Millipore, Upstate Biotechnology, catalog number: 05-499 )
- Anti-alpha-tubulin antibody (generously provided by Dr. A. Baines)
Note: Commercially available antibodies against alpha-tubulin (such as Abcam, catalog number: ab6161 ) can be used.
- Goat HRP conjugated-anti-mouse antibody (Life Technologies, Biosource, catalog number: A10551 )
- YES liquid medium (see Recipes)
- STOP buffer (see Recipes)
- PEMS buffer (see Recipes)
- 2x HBS buffer (see Recipes)
- Lysis buffer (see Recipes)
- 2x Laemmli protein sample buffer (see Recipes)
Equipment
- Air- (Tykyo Rikakikai, Eyela, model: FMC-1000 ) or water bath- (Taitec, model: MM-10 ) incubator shaker
- Centrifuges equipped with 50 ml tubes (Tomy Digital Biology, model: AX-501 ) and 1.5 ml microtubes (Tomy Digital Biology, model: MX-107 )
- Heat block (TAITEC, model: DTU-1BN) or water bath (Taitec, model: EXN-B )
- Light microscope (Nikon Corporation, model: Eclipse E200 )
- Electrophoresis apparatus (Bio-Rad Laboratories, catalog numbers: 165-8002JA and 164-5052 )
- Transfer unit (Bio-Rad Laboratories, catalog numbers: 170-3930JA and 170-3935JA )
Procedure
- Culture fission yeast cells in 200 ml of YES medium in a 500 ml flask at 26 °C.
Note: Stop culturing during the mid-log phase (OD595 = ~1.0).
- Prepare approximately 2.5 x 108 cells by dilution of inoculum to 25 ml at OD595 = 0.5.
- Harvest the cells by centrifugation at 400 x g for 5 min at 4 °C.
- Resuspend the cell pellet in 1 ml of ice-cold STOP buffer and transfer the cell suspension to a 1.5 ml microtube.
- Centrifuge the cell suspension at 400 x g for 5 min at 4 °C and pour out the supernatant (STOP buffer) by aspiration.
- Place the microtube containing the cell pellet on ice for 5 min.
- Resuspend the cell pellet in 1 ml of PEMS buffer dissolving 1 mg/ml lysing enzymes and 1 mg/ml Zymolyase 100T.
- Incubate the cell suspension for 20 min at 37 °C for making spheroplasts which readily burst for enabling to fractionate chromatin-bound and -unbound proteins at the later step.
Note: To test whether these lysing enzymes work adequately, take a small volume of the cell suspension, add the equal volume of 10% SDS, and then monitor yeast lysis progression by light microscopy. If lysing is successful, almost all cells (spheroplasts) are expected to burst in the presence of SDS.
- Centrifuge the spheroplast suspension at 400 x g for 5 min at 4 °C. Pour out the supernatant (PEMS buffer) carefully by aspiration.
- Wash the spheroplast pellet twice by resuspending in 1 ml of 1.2 M sorbitol to avoid bursting of spheroplasts, followed by centrifugation at 400 x g for 5 min at 4 °C.
- Lyse the spheroplasts by resuspending in 0.5 ml of ice-cold lysis buffer, followed by incubation on ice for 5 min.
- Take 1/10 aliquot of the resulting lysate and save it as the whole cell extract (optional).
Note: To check the protein expression of our interest, we recommend an immunoblot of the whole cell extract with the other samples.
- Centrifuge the rest of the lysate at 22,000 x g for 15 min at 4 °C.
- Save the supernatant as the chromatin-unbound fraction.
- Wash the pellets twice by resuspending in 1 ml of lysis buffer, followed by centrifugation at 22,000 x g for 5 min at 4 °C.
- Resuspend the pellet in 0.45 ml of lysis buffer and save it as the chromatin-bound fraction.
- Add an equal volume of 2x Laemmli protein sample buffer to all of the saved fractions.
- Apply an equal volume of these fractions to the 8% SDS-PAGE gel and immunoblot using antibodies against histone H3 (1/5,000 dilution), alpha-tubulin (1/5,000 dilution) and specific protein(s).
Note: We detected the chromatin association of Pcf1-GFP protein by using the anti-GFP antibody (1/1,000 dilution). The secondary antibody was diluted at 1/10,000.
Representative data
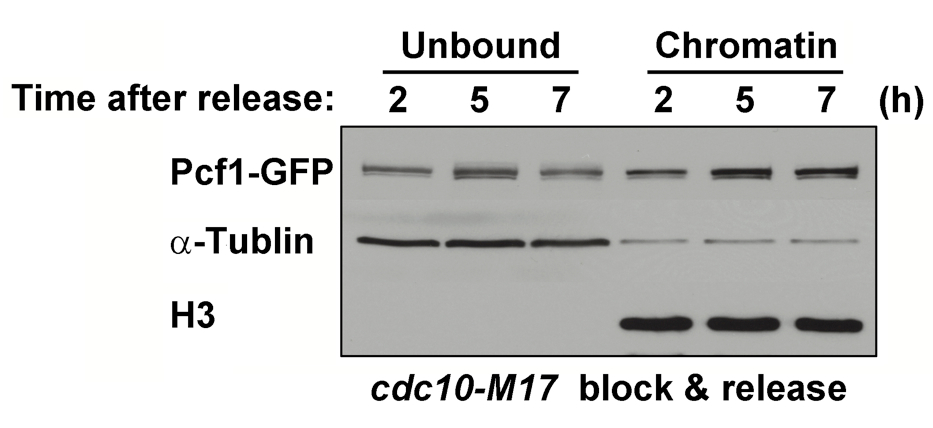
Figure 1. The cdc10-M17 mutant cells were grown in YES medium at 26 °C, synchronized to G1 phase by incubation for 4 h at 36 °C, and returned to 26 °C. Released cells into S phase were harvested at indicated time(s) and subjected to chromatin fractionation assay to monitor the chromatin association of Pcf1-GFP.
Notes
- The proper performance of the assay can be easily confirmed by immunoblots using anti-histone H3 and anti-alpha-tubulin antibodies, since histone H3 and alpha-tubulin are fractionated to the chromatin-bound and -unbound fractions, respectively. In some cases as shown in Figure 1, alpha-tubulin was fractionated into the chromatin-bound fraction. This contamination was probably due to low efficiency of cell lysis or insufficient washout of the chromatin-unbound proteins. Therefore, we recommend the reader(s) to check the former possibility by monitoring yeast lysis progression in the presence of SDS and adjusting cell number, concentration of lysing enzymes, and/or reaction time, if necessary. For the latter possibility, additional wash of the pellets should be done to remove the chromatin-unbound proteins completely.
Recipes
- YES liquid medium
5 g/L Bacto yeast extract
30 g/L glucose
225 mg/L adenine, histidine, leucine, uracil and lysine hydrochloride
- STOP buffer
150 mM NaCl
50 mM NaF
10 mM EDTA
1 mM NaN3
pH 8.0
- PEMS buffer
100 mM PIPES
50 mM EDTA
10 mM MgSO4
1.2 M sorbitol
pH 6.9
- 2x HBS buffer
50 mM MOPS
120 mM beta-glycerophosphate
30 mM MgCl2
30 mM EGTA
30 mM p-nitrophenylphosphate
0.2 mM Na3VO4
pH 7.2
- Lysis buffer
2x HBS buffer 5 ml
10 % Triton X-100 0.5 ml
2 M sorbitol 2 ml
100 mM dithiothreitol (DTT) 0.1 ml
0.1 mM phenylmethanesulfonyl fluoride (PMSF) 0.1 ml
Complete mini 1 tablet
Fill up to 10 ml with distilled water
- 2x Laemmli protein sample buffer
0.1 M Tris-HCl
4 % sodium dodecyl sulfate (SDS)
20 % glycerol
0.2 % bromophenolblue (BPB)
20 % 2-mercaptoethanol (2-ME)
pH 6.8
Acknowledgments
This protocol was described in our previous report (Kunoh and Habu, 2014). We express our sincere appreciation to Dr. Mahito Sadaie (Graduate School of Biostudies, Kyoto University) for his technical advice on this protocol. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan to T.H. and JST-CREST to Professor Jun Takada, Okayama University.
References
- Kunoh, T. and Habu, T. (2014). Pcf1, a large subunit of CAF-1, required for maintenance of checkpoint kinase Cds1 activity. Springerplus 3: 30.
- Ogawa, Y., Takahashi, T. and Masukata, H. (1999). Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol Cell Biol 19(10): 7228-7236.
- Sadaie, M., Kawaguchi, R., Ohtani, Y., Arisaka, F., Tanaka, K., Shirahige, K. and Nakayama, J.. (2008). Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol Cell Biol 28(23): 6973-6988.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kunoh, T. and Habu, T. (2014). Chromatin Fractionation Assay in Fission Yeast. Bio-protocol 4(14): e1185. DOI: 10.21769/BioProtoc.1185.
Category
Microbiology > Microbial genetics > DNA > Chromosomal
Molecular Biology > DNA > DNA-protein interaction
Molecular Biology > DNA > DNA damage and repair
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









