- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Hedgehog (Hh) Reporter Activity Assay
Published: Vol 4, Iss 14, Jul 20, 2014 DOI: 10.21769/BioProtoc.1182 Views: 15756
Reviewed by: Hong Lok LungFanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Organotypic Slice Culture of the Embryonic Mouse Brain
James M. Clegg and Thomas Pratt
Jul 5, 2020 5892 Views

Generation of Mouse Primitive Endoderm Stem Cells
Yasuhide Ohinata [...] Haruhiko Koseki
Nov 20, 2023 2452 Views

An Efficient Method for Immortalizing Mouse Embryonic Fibroblasts by CRISPR-mediated Deletion of the Tp53 Gene
Srisathya Srinivasan and Hsin-Yi Henry Ho
Jan 20, 2025 2758 Views
Abstract
This protocol is for testing responses of a candidate cell line/cell lines to Hh ligands or Hh pathway agonists stimulation. This protocol can also be adapted to screen small molecule libraries or biologics that contain activities to either increase or decrease Hh pathway responses. Canonical Hh signaling activity transcriptionally induces Hh target genes that contain consensus Gli binding element. Hh-responsive cells transiently or stably expressing luciferase protein under the regulation of the Gli promoter element can be used to report stimulus-dependent Hh-pathway activity.
Materials and Reagents
- NIH3T3 cells or NIH3T3-Light2 cells (ATCC, catalog number: JHU-68 )
Note: NIH3T3-Light2 cells were initially generated in Dr. Beachy’s Laboratory and were deposited in ATCC, the public available source of biomedical reagents including laboratory made cell lines. This cell line is available from Dr. Beachy’s lab per request. - Hh-conditioned medium
- Fetal bovine serum (FBS)
- Bovine calf serum (BCS)
- DMEM (high glucose) (Life Technologies, InvitrogenTM, catalog number: 11965 )
- TransIT-2020 (Mirius, catalog number: MIR5404 )
- 8XGli-Firefly luciferase expression construct
Note: The 8XGli-Firefly luciferase expression construct is made at Dr. Beachy’s Laboratory. Eight consecutive repeats of Gli responding element (8XGli) are PCR amplified and cloned into the minimal enhancer region of pGL3-Luciferase [promega] vector. Map of pGL3 is available from promega. - pRL-SV40-Renilla luciferase expression construct
Note: The pRL-SV40-Renilla luciferase expression construct is made at Dr. Beachy’s Laboratory. - Dual-Luciferase @ Reporter Assay System (Promega Corporation, catalog number: E1960 )
- Growth media (see Recipes)
- Shh-conditioned media (see Recipes)
- Serum deprived media (see Recipes)
Equipment
- 15 ml thermal scientific Nunc conical tubes
- T75 culture flask (BD Biosciences, Falcon®, catalog number: 353135 )
- 24-well tissue culture plate (BD Biosciences, Falcon®, catalog number: 353047 )
- 96-well flat-bottom plate (Corning, Costar® , catalog number: 3915 )
- Centrifuge (Eppendorf, model: 8810R )
- Water bath
- CO2 incubator
- Berthold Luminometer (Berthold Technologies, model: Centro XS LB960 )
Procedure
- Grow low passage NIH3T3 or NIH3T3-light2 cells from liquid nitrogen stock
- Briefly thaw cells at 37 °C water bath.
- Pipette cells into 15 ml thermal scientific Nunc conical tubes and spin for 5 min at 1,500 x g. Aspirate media, re-suspend cells in 15 ml growth media and plate in T75 culture flask. (Cells are plated at the density at 1 x 106 and are cultured at 37 °C with 5% CO2.)
- Split the cells when they grow to 60% sub-confluency.
Note: Never let cells reach 100% confluency. - The cell line should be subcultured for one or two time before performing the actual assay. Then plate the cells into 24-well plates as triplicate according to the experimental conditions (for NIH3T3 cells, plate as 5 x 104 cells/well; For NIH3T3-Light2 cells, plate as 4 x 105 cells/well).
- Briefly thaw cells at 37 °C water bath.
- Transfection (only for NIH3T3 cells, as NIH3T3-Light stably express dual luciferase reporter constructs, if work with NIH3T3-Light2 cells, go directly to step B3).
- Day 0: Seed cells (density as above-mentioned) in a total volume of 500 μl complete growth media (DMEM/10%FBS).
- Day 1: Transfection (use ratio of 8XGli firefly luciferase: pRL-SV40-Renilla luciferase as 10:1; use 1.5 μl TransIT-2020/300 ng of DNA/well).
- Day 2: Change media 24 h after transfection (optimal).
- Day 3: Change to serum deprived media.
- Day 5: Lyse cells (1x whole cell lysis buffer available from the Dual-Luciferase @ Reporter Assay System) and collect aliquot of supernatant (20 μl) from cell lysis, plate them into 96-well plate.
- Day 0: Seed cells (density as above-mentioned) in a total volume of 500 μl complete growth media (DMEM/10%FBS).
- Reading luminescence signal
- Thaw dual-luciferase reporter reagents.
- Flash and prime Berthold luminometer with firefly luciferase and renilla luciferase substrate reagents. Firefly luciferase substrate is a derivative of D-luciferin. In response to D-luciferin enzymatic activity (firefly luciferase), a firefly luciferase chemical reaction will generate oxyluciferin and a specific light signal at 560 nm that can be detected by the luminometer. Renilla luciferase substrate is a derivative of Coelenterazine. In response to renilla enzymatic activity (renilla luciferase), a renilla luciferase chemical reaction will generate Coelenteramide and a specific light signal at 480 nm that can be detected by the luminometer. 20 μl of each substrate is added sequencially by the luminometer and light signals generated are instantly measured by the luminometer (light signals are detected at an enclosed chamber of the luminometer at ambient temperature).
- Read firefly and renilla luciferase signals (firefly luciferase signal is detected at 560 nm and renilla luciferase signal is detected at 480 nm).
- Thaw dual-luciferase reporter reagents.
Representative data
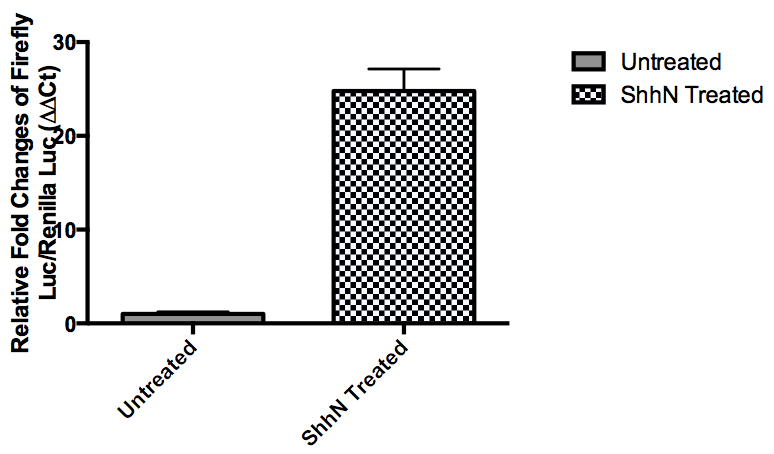
Figure 1. Firefly and renilla luciferase signals are measured at untreated and ShhN-treated conditions. Relative firefly/renilla signals are normalized as fold of induction to untreated conditions.
Note: Relative Firefly Luciferase (FL)/Renilla Luciferase(RL) = Raw FL/Raw RL. Fold changes of FL/RL at Shh-stimulated condition is normalized to FL/RL at unstimulated condition.
Recipes
- Growth media
For NIH3T3 cells: DMEM + 10% FBS + 1% PS
For NIH3T3-light2 cells: DMEM + 10% BCS + 1% PS - Shh-conditioned media
Note: Shh-conditioned media is prepared by collecting supernatant of HEK293 cells stably expressing N-terminal truncated form of Shh protein.
Shh-conditioned media is diluted in a ratio of 1:10 in DMEM + 2% FBS + 1% PS. - Serum deprived media
DMEM
2% FBS containing various concentrations of Hh agonists or diluted Shh-conditioned media
Acknowledgments
This protocol is adapted from published work (Kim et al., 2010). I thank the current and past members of the Beachy lab, Stanford University, who contributed to the development of this protocol. I acknowledge the Susan G. Komen for the Cure Postdoctoral Fellowship: KG111253.
References
- Kim, J., Lee, J. J., Kim, J., Gardner, D. and Beachy, P. A. (2010). Arsenic antagonizes the Hedgehog pathway by preventing ciliary accumulation and reducing stability of the Gli2 transcriptional effector. Proc Natl Acad Sci U S A 107(30): 13432-13437.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zhao, C. (2014). Hedgehog (Hh) Reporter Activity Assay. Bio-protocol 4(14): e1182. DOI: 10.21769/BioProtoc.1182.
Category
Developmental Biology > Cell signaling > Ligand
Stem Cell > Embryonic stem cell > Maintenance and differentiation
Cell Biology > Cell signaling > Development
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









