- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
ChIP-Seq in Candida albicans
Published: Vol 4, Iss 12, Jun 20, 2014 DOI: 10.21769/BioProtoc.1158 Views: 19188
Reviewed by: Fanglian HeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Chromatin Immunoprecipitation (ChIP) Assay for Detecting Direct and Indirect Protein – DNA Interactions in Magnaporthe oryzae
Gang Li [...] Richard A. Wilson
Nov 5, 2015 15992 Views

Biochemical Analysis of Dimethyl Suberimidate-crosslinked Yeast Nucleosomes
Yuichi Ichikawa and Paul D. Kaufman
Mar 20, 2018 7865 Views

Identifying Protein Interactions with Histone Peptides Using Bio-layer Interferometry
Bingbing Ren [...] Ee Sin Chen
Sep 20, 2018 8915 Views
Abstract
Systems biology approaches can be used to study the regulatory interactions occurring between many components of the biological system at the whole-genome level and decipher the circuitries implicated in the regulation of cellular processes, including those imparting virulence to opportunistic fungi. Candida albicans (C. albicans) is a leading human fungal pathogen. It undergoes morphological switching between a budding yeast form and an elongated multicellular hyphal form. This transition is required for C. albicans’ ability to cause disease and is regulated through highly interconnected regulatory interactions between transcription factors (TFs) and target genes. The chromatin immunoprecipitation (ChIP)-High-throughput sequencing (Seq) technology (ChIP-Seq) is a powerful approach for decoding transcriptional regulatory networks. This protocol was optimized for the preparation of ChIP DNA from filamenting C. albicans cells followed by high-throughput sequencing to identify the targets of TFs that regulate the yeast-to-hyphae transition.
Materials and Reagents
- C. albicans strains expressing or not a functional epitope-tagged transcription factor grown under filamentation-inducing conditions in liquid medium [50 ml, e.g. Lee’s medium at 37 °C (Lee et al., 1075)]
- 37% formaldehyde (Sigma-Aldrich, catalog number: F8775 )
- Liquid nitrogen
- Dynabeads® Pan Mouse IgG (5 ml) (Life Technologies, catalog number: 11041 )
- Appropriate mouse monoclonal antibody directed against the epitope tag fused to TF [e.g. mouse monoclonal anti-HA antibody, HA-probe Antibody (F-7), Santa Cruz, catalog number: sc-7392 ]
- 100 mM Phenylmethylsulfonyl fluoride (PMSF) dissolved in isopropanol (100x stock solution)
- DNase-free RNase A solution (10 mg/ml) (Thermo Fisher Scientific, catalog number: EN0531 )
- Bovine Serum Albumin (BSA) (Sigma-Aldrich, catalog number: B4287 )
- Proteinase K solution (20 mg/ml) (Life Technologies, catalog number: AM2546 )
- Glycogen (20 mg/ml) (Thermo Fisher Scientific, catalog number: FERR0561 )
- Phenol: Chloroform: Isoamyl alcohol (25: 24: 1) (Sigma-Aldrich, catalog number: P2069 )
- 5 M NaCl solution
- 70%, 100% freezer-cold Ethanol
- Quant-iTTM PicoGreen® dsDNA Assay Kit (Life Technologies, catalog number: P11496 )
- TruSeqTM DNA Sample Preparation Kit v.2 (Illumina, catalog numbers: FC-121-2001 , FC-121-2002 )
- TruSeqTM DNA Sample Preparation Guide v.2 (Illumina, catalog number: FC-930-1021)
- QIAquick PCR Purification Kit (QIAGEN, catalog number: 28104 )
- MinElute PCR Purification Kit (QIAGEN, catalog number: 28004 )
- E-Gel® iBaseTM and E-Gel® Safe ImagerTM Combo Kit (Life Technologies, catalog number: G6465 )
- 2% E-Gel® SizeSelectTM Agarose Gels (Life Technologies, catalog number: G6610-02 )
- Glycine (Sigma-Aldrich, catalog number: 50046 )
- SDS (Sigma-aldrich, catalog number: L3771 )
- 2.5 M glycine (see Recipes)
- 10% SDS (see Recipes)
- Lee’s medium (see Recipes)
- TBS buffer (see Recipes)
- Lysis buffer (see Recipes)
- PBS/0.5% BSA (see Recipes)
- Wash buffer (see Recipes)
- Protease inhibitor cocktail tablets (Roche Diagnostics, catalog number: 11697498001 ) (see Recipes)
- TE solution (see Recipes)
- TE/SDS solution (see Recipes)
Equipment
- 15 ml, 50 ml Falcon tubes
- Racks, including a 50-ml Falcon tube rack (Unwire test tube rack, for 30 mm tubes; holds 24) (Thermo Fisher Scientific, Nalgene®, catalog number: 14-809-30 )
- VSR-50 laboratory platform rocker (Pro Scientific, catalog number: PSI-512000-00 ) or equivalent
- Waste container
- 1.5 ml screw-cap tubes
- 0.5 ml PCR tube
- FastPrep®-24 instrument (MP Biomedicals, catalog number: 116004500 )
- Microscope (e.g. Zeiss Axiostar Plus)
- 18 G x 1½ inch needles (BD Biosciences, catalog number: 305196 )
- 2 ml Eppendorf conical tubes (Eppendorf, catalog number: 022363352 )
- Probe sonicator (e.g. MSE Soniprep 150 Plus, exponential microprobe, end diameter 3 mm) [MSE (UK), catalog number: 38121-114A ]
- Glass beads (0.5 mm diameter) (Bio Spec Products, catalog number: 11079105 )
- Hematology/chemistry mixer 346 (Thermo Fisher Scientific, catalog number: 14-059-346 )
- DynaMag Spin magnet system (Life Technologies, model: 123-20D )
- Fluorescence reader (e.g. Tecan Trading AG, Infinite®, model: M200 )
- BioAnalyzer 2100 (Agilent)
- HiSeq 2000 sequencer (Illumina)
Software
- Galaxy NGS data analysis software (https://main.g2.bx.psu.edu/)
- ChIP-Seq (MACS) peak-finding algorithm software (http://liulab.dfci.harvard.edu/MACS/)
Procedure
- Protein-DNA crosslinking
- Start a preculture of both tagged and untagged (control) C. albicans strains in 10 ml of rich medium (e.g. Yeast Peptone Dextrose) - allow to grow overnight under vigorous shaking (200 rpm) at 30 °C.
- Dilute cells to an optical density at 600 nm (OD600nm) of 0.4 in 50 ml of an appropriate filamentation-inducing liquid medium (e.g. Lee’s medium at 37 °C) and allow to grow under vigorous shaking (200 rpm) until cells reach the equivalent of OD600nm = 1.0 (or typically during 4 h in Lee’s medium at 37 °C).
- Add 1.4 ml of 37% formaldehyde to a 50 ml Falcon tube and transfer the culture (48.6 ml) to the Falcon tube. Screw the caps tightly to prevent leakage of formaldehyde-containing liquid.
- Place the Falcon tubes in a suitable rack and tape them. Place the rack on the rocker and agitate during 30 min at room temperature (agitation at 45 rpm using the VSR-50 laboratory platform rocker).
- Add 2.5 ml of 2.5 M glycine to stop the cross-linking reaction and incubate 10 min with agitation on the rocker.
- Centrifuge the samples for 5 min at 3,500 rpm (2,465 x g). Discard the supernatant (containing formaldehyde) in a suitable waste container. Wash the cells 3 times with 10 ml TBS buffer, by agitating, quickly (1-2 min) centrifuging at 3,500 rpm (2,465 x g) and resuspending in 10 ml TBS buffer.
- Centrifuge the samples for 5 min at 3,500 rpm (2,465 x g) and discard the supernatant. Using the remaining liquid, resuspend the cell pellets and transfer to a 1.5 ml screw-cap tube.
- Briefly centrifuge the samples and remove the remaining supernatant. Snap-freeze samples in liquid nitrogen. Store tubes at -80 °C until use.
- Start a preculture of both tagged and untagged (control) C. albicans strains in 10 ml of rich medium (e.g. Yeast Peptone Dextrose) - allow to grow overnight under vigorous shaking (200 rpm) at 30 °C.
- Preparing Dynabeads for coupling to antibody (work on ice or in a 4 °C room)
- Coupling of Dynabeads to antibody for immunoprecipitation (IP) is made the day preceding the scheduled day for preparing extracts. The main stock of beads is vortexed to resuspend the beads and 50 µl of beads per IP are used.
- Transfer the required volume of beads to a 15 ml Flacon tube (e.g. for 6 samples, transfer ~300 µl).
- Centrifuge quickly and remove the supernatant.
- Wash the beads with 5 ml PBS/0.5% BSA (make a fresh solution, e.g. 0.25 g BSA in 50 ml PBS).
- Typically 2 µg of antibody per IP are needed for coupling to the beads at a dilution of 1/10. Resuspend the beads in appropriate volume of PBS/0.5% BSA and add the required volume of antibody at a 1/10 dilution (e.g. for 6 samples and an antibody stock solution at 0.2 µg/µl, transfer 60 µl of antibody to 600 µl of beads resuspended in PBS/0.5% BSA).
- Incubate the beads-antibody overnight at 4 °C on a hematology/chemistry mixer.
- Quickly centrifuge the 15-ml Falcon tube. Wash the beads-antibody with PBS/BSA 0.5% and resuspend in 30 µl lysis buffer per IP (for 6 samples, resuspend in ~180 µl of lysis buffer).
- Transfer the required volume of beads to a 15 ml Flacon tube (e.g. for 6 samples, transfer ~300 µl).
- Coupling of Dynabeads to antibody for immunoprecipitation (IP) is made the day preceding the scheduled day for preparing extracts. The main stock of beads is vortexed to resuspend the beads and 50 µl of beads per IP are used.
- Preparing total extracts and DNA shearing (work on ice or in a 4 °C room)
- Thaw the cell pellets from step 8 on ice and resuspend them in 700 µl of lysis buffer supplemented with protease inhibitor cocktail and 1 mM PMSF (1x final each).
- Add the equivalent of 0.5 ml PCR tube of glass beads.
- Prepare total cell extracts by bead beating using a FastPrep-24 instrument with 6 runs during 1 min each at 6.0 m/sec and 1 min on ice in between (These settings led to efficient breakage of hyphal cells.).
- Make sure of efficient cell breakage by examining 5 µl of the FastPrep processed sample under a light microscope.
- Punch a hole at the bottom of the screw cap tube containing the total extracts + beads using an 18 G x 1½ inch needle and quickly place it on a clean 2 ml conical tube. The bottom part of the 1.5 ml screw-cap tube carrying the beads + total extracts has to be introduced into the 2 ml microtube.
- Quickly centrifuge (spinning pulse from 0 to ~9,000 rpm) the assembled tubes to collect the total extract from the 1.5 screw cap tube in the 2 ml collection tube. Make sure there are no remaining cells adhered to the glass beads in the 1.5 ml screw cap tube.
- Resuspend the collected cell debris + extracts in the 2 ml collection tube and transfer to a clean 1.5 ml microtube.
- Sonicate the extracts 4 times during 20 sec at power 8 (knob position) for an output signal amplitude of 21 (Microns, Peak to Peak) using a probe sonicator. Try to get ~200 to 300 bp DNA fragments on average (Figure 1).
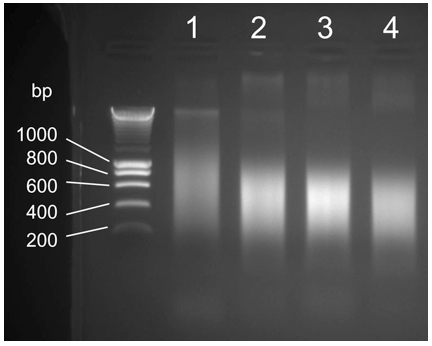
Figure 1. Optimization of DNA shearing by sonication. Total extracts from step 16 (~700 µl) are sonicated 4 times during 20 sec each with 1 min incubation on ice in-between. 4 different sonication power settings [top; 1, knob position 2 output power 6 (Micron, Peak to Peak); 2, knob position 4 output power 10.5; 3, knob position 6 output power 15; 4, knob position 8 output power 21] are tested to optimize DNA fragment size range. Condition 4 is the optimal DNA fragment size range for high-throughput sequencing (~100 bp to maximum 500 bp, maximum average size ~300 bp) DNA ladder sizes are indicated at the left of the panel (bp, base pairs).
- Centrifuge the sonicated samples at 14,000 rpm during 10 min at 4 °C and transfer the supernatant (This is the whole cell extract, WCE.) to a clean 1.5 ml microtube.
- Thaw the cell pellets from step 8 on ice and resuspend them in 700 µl of lysis buffer supplemented with protease inhibitor cocktail and 1 mM PMSF (1x final each).
- Chromatin immunoprecipitation (work on ice or in a 4 °C room)
- Transfer 500 µl of the WCE to a new 1.5 ml microtube and add 30 µl of Dynabeads coupled to antibody from step B9f.
- Incubate the Dynabeads-antibody-WCE mixture overnight at 4 °C under rotation on a hematology/chemistry mixer.
- Transfer 500 µl of the WCE to a new 1.5 ml microtube and add 30 µl of Dynabeads coupled to antibody from step B9f.
- Washing and reversing crosslinks
- In a cold room (4 °C), place the samples on the DynaMag Spin magnet system to allow separation of the Dynabeads from the WCE. Wash twice with 1 ml lysis buffer, twice with 1 ml lysis buffer supplemented with 360 mM NaCl, twice with 1 ml wash buffer and once with 1 ml TE buffer. For each washing step, put the tubes on the magnet, rotate the magnet + tubes 5-6 times to allow efficient attraction of beads, discard the supernatant, add 1 ml of respective buffer, remove the magnet and vigorously agitate the tubes 30-50 times.
- Centrifuge during 2 min at 4,000 rpm at 4 °C and remove the supernatant.
- Add 100 µl of TE/SDS solution to the bead pellet. Vortex gently (medium speed) to resuspend and incubate overnight at 65 °C. Vortex for 1 min before leaving at night and vortex again the following morning. Incubate for another 30 min at 65 °C before proceeding.
- In a cold room (4 °C), place the samples on the DynaMag Spin magnet system to allow separation of the Dynabeads from the WCE. Wash twice with 1 ml lysis buffer, twice with 1 ml lysis buffer supplemented with 360 mM NaCl, twice with 1 ml wash buffer and once with 1 ml TE buffer. For each washing step, put the tubes on the magnet, rotate the magnet + tubes 5-6 times to allow efficient attraction of beads, discard the supernatant, add 1 ml of respective buffer, remove the magnet and vigorously agitate the tubes 30-50 times.
- DNA purification
- Vortex again at medium speed and incubate during 30 min at 65 °C.
- Centrifuge at 14,000 rpm during 2 min and transfer the supernatant to a new 1.5 ml microtube.
- Add a mixture of 295 µl TE, 3 µl of RNase A (stock at 10 mg/ml) and 2 µl of glycogen (from stock at 20 mg/ml). Vortex to mix the samples (400 µl final) and incubate at 37 °C during 2 h.
- Add 15 µl of 10% SDS and 7.5 µl of proteinase K solution. Vortex gently and incubate 2 h at 37 °C.
- Extract twice with 400 µl phenol:chloroform:isoamyl alcohol (25:24:1) by vortexing for 1 min and centrifuging at 14,000 rpm during 10 min at room temperature. Transfer the supernatant from each extraction step to a clean 1.5 ml microtube.
- Add 16 µl of 5 M NaCl to the extracted supernatant from step F28 (should be ~350 µl). Vortex then add 2.5 volumes of freezer-cold ethanol (~875 µl). Stored overnight at -20 °C.
- Centrifuge at 14,000 rpm during 40 min at 4 °C.
- Remove the supernatant and wash with 1 ml of freezer-cold 70% ethanol.
- Resuspend the pellet in a convenient volume of H2O (typically 50 µl).
- Quantify the IP DNA using the Quant-iTTM PicoGreen® dsDNA Assay Kit following the manufacturer’s instructions.
- Vortex again at medium speed and incubate during 30 min at 65 °C.
- DNA library generation and high-throughput sequencing
- Use 10 ng of IP DNA for library generation. Generate the Illumina library using the TruSeq DNA sample preparation kit v.2 as recommended by the TruSeq DNA sample preparation v.2 guide. Steps include (with some modifications):
- End repair as recommended in TruSeq DNA sample preparation v.2 guide (page 43).
- Purification of end-repaired DNA with QIAquick PCR Purification kit as recommended by the manufacturer, instead of using Agencourt AMPure XP beads.
- Adenylation of 3’ DNA ends as recommended in TruSeq DNA sample preparation v.2 guide (page 47).
- Ligation of indexed adapters as recommended in TruSeq DNA sample preparation v.2 guide (page 49).
- Clean up with Qiagen MinElute PCR purification kit as recommended by the manufacturer, instead of using Agencourt AMPure XP beads.
- Use E-Gel® iBaseTM and E-Gel® Safe ImagerTM Combo Kit and E-Gel® SizeSelectTM Agarose Gels, 2% following the instructions provided by the manufacturer to purify the desired library fragments, instead of using 2% agarose with SyBr Gold gel. Migrate samples on E-gel during 20 min as recommended by Invitrogen. Select fragments with an average size of ~300 bp.
- Enrich DNA fragments by amplification with PCR for 10 cycles as recommended in TruSeq DNA sample preparation v.2 guide (page 58).
- Clean up PCR with QIAquick PCR Purification kit as recommended by the manufacturer, instead of using Agencourt AMPure XP beads.
- Validate library and perform quality control by loading and migrating DNA on a BioAnalyzer 2100 device with a High Sensitivity DNA chip (Figure 2) following the manufacturer’s instructions and as indicated on page 62 of the TruSeq DNA sample preparation v.2 guide.
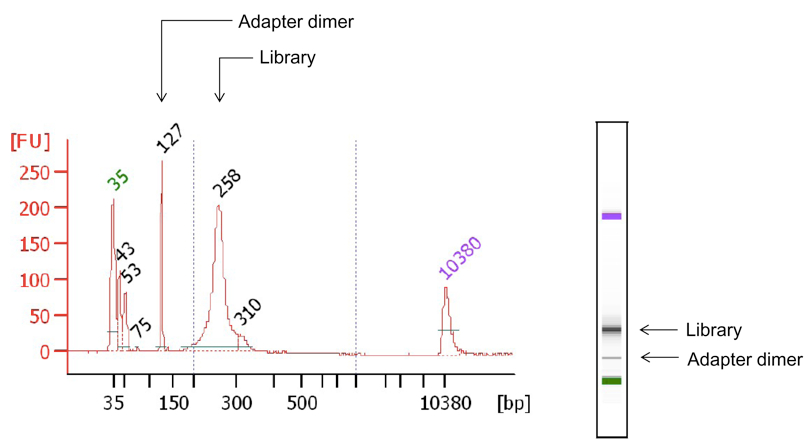
Figure 2. Quality control of purified Illumina adapter-ligated DNA library using the BioAnalyzer 2100. Representative BioAnalyzer 2100 migration profile of Illumina high-throughput sequencing DNA library from ChIP assay (Library). Illumina adapter dimers are indicated (Adapter dimer). DNA fragment sizes are indicated on the x-axis [(bp), base pairs] and DNA abundance on the y-axis [(FU), fluorescence units].
- End repair as recommended in TruSeq DNA sample preparation v.2 guide (page 43).
- Dilute ChIP library samples to 2 nM, denature DNA samples and load onto an Illumina HiSeq 2000 sequencer flow cell lane for single-read (51 base pairs per read) high-throughput sequencing following the manufacturer’s instructions.
- The resulting read files (FASTQ) are pre-processed with the online Galaxy NGS data analysis software (Blankenberg et al., 2010; Giardine et al., 2005). Steps include:
- Quality control analyses of the FASTQ files using FastQC version 0.52 on the Galaxy web interface - proceed to clipping of adapter-contaminated sequences using Clip version 1.0.1.
- Mapping of the resulting files to the C. albicans assembly 21 genome (http://www.candidagenome.org/download/chromosomal_feature_files/C_albicans_SC5314/) using the Bowtie algorithm (Galaxy: Click on NGS mapping>Map with Bowtie for Illumina version 1.1.2) - the Assembly 21 of the C. albicans genome can be imported into Galaxy by clicking on User>Custom builds (top window of the Galaxy web interface). Mapping will generate BAM files for processing with peak finding algorithms.
- Quality control analyses of the FASTQ files using FastQC version 0.52 on the Galaxy web interface - proceed to clipping of adapter-contaminated sequences using Clip version 1.0.1.
- Proceed to peak finding with the generated BAM files using the Model-Based Analysis for ChIP-Seq (MACS) peak-finding algorithm software and following the instructions found on one of the two corresponding detailed protocols (Feng et al., 2012; Feng et al., 2011).
- Use 10 ng of IP DNA for library generation. Generate the Illumina library using the TruSeq DNA sample preparation kit v.2 as recommended by the TruSeq DNA sample preparation v.2 guide. Steps include (with some modifications):
Recipes
- 2.5 M glycine
Dissolve 93.8 g of glycine in 500 ml H2O
- 10% SDS
Dissolve 10 g of SDS in 100 ml H2O
- Lee’s medium (for 1 L)
5.0 g of (NH4)2SO4
0.2 g of MgSO4.7H2O
2.5 g of K2HPO4
5.0 g of NaCl
12.5 g of D-Glucose
0.5 g of L-Alanine
1.3 g of L-Leucine
1.0 g of L-Lysine
0.1 g of L-Methionine
0.0714 g of L-Ornithine
0.5 g of L-Phenylalanine
0.5 g of L-Proline
0.5 g of L-Threonine
0.001 g of Biotin
Distilled water to 1 L
Combine ingredients (except biotin) and autoclave at 110 °C for 20 min
Add biotin
pH 6.8 ± 0.05
- 5x TBS buffer (for 1 L)
100 ml of 1M Tris-HCl (pH 7.5) (100 mM final at 5x)
150 ml of 5M NaCl (750 mM NaCl final at 5x)
H2O to 1 L (750 ml)
Filter sterilize
Stored at 4 °C
- 1x lysis buffer (for 500 ml)
25 ml of 1M HEPES-KOH (pH 7.5) (50 mM final at 1x)
14 ml of 5M NaCl (140 mM final at 1x)
1 ml of 500 mM EDTA (1 mM final at 1x)
50 ml of 10% Triton X100 (1% final at 1x)
10 ml of 5% Na-deoxycholate (0.1% final at 1x)
H2O to 500 ml (400 ml)
Filter sterilize
Stored at 4 °C
- 1x PBS/0.5% BSA (for 50 ml)
Preparation of 1x PBS solution
1 L at 1x: To ~800 ml of H2O add 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, 0.24 g of KH2PO4 and stirr with a magnetic stirrer. Adjust the pH to 7.4 with HCl and complete with H2O to 1 L. Stored at room temperature.
Preparation of PBS/0.5% BSA
Add 0.25 g of Bovine Serum Albumin to 50 ml 1x PBS in a 50-ml Falcon tube. Do not mix, simply stored at 4 °C for ~ 30 min to 1 h.
- 1x wash buffer (for 500 ml)
5 ml of 1 M Tris-HCl (pH 8.0) (10 mM final at 1x)
25 ml of 5 M LiCl (250 mM final at 1x)
2.5 ml of 100% NP40 (0.5% final at 1x)
25 ml of 10% Na-deoxycholate (0.5% final at 1x)
1 ml of 500 mM EDTA (1mM final at 1x)
H2O to 500 ml (441.5 ml)
- Protease inhibitor cocktail tablets
25x stock solution made by dissolving one tablet in 2 ml H2O
Aliquot 100 µl in 1.5 ml tubes
Stored at -20 °C
- TE solution
10 mM Tris (pH 8.0)
1 mM EDTA
- TE/SDS solution
10 mM Tris (pH 8.0)
1 mM EDTA
1% sodium dodecyl sulfate
Acknowledgments
This protocol was adapted from Drouin & Robert « Genome-wide Location Analysis of Chromatin-associated Proteins by ChIP on CHIP: Controls Matter » available at : http://www.ircm.qc.ca/LARECHERCHE/axes/Biologie/Chromatine/Documents/ProtocoleIRCM_LevureYeast1.pdf and Znaidi et al. (2013). This work was supported by grants from the European commission (FinSysB PITN-GA-2008-214004), the Agence Nationale de la Recherche (KANJI, ANR-08-MIE- 033-01) and a BIOASTER-Sanofi-Alliance pour les Sciences de la Vie et de la Santé (AVIESAN) joint program to Dr. Christophe d’Enfert.
References
- Blankenberg, D., Von Kuster, G., Coraor, N., Ananda, G., Lazarus, R., Mangan, M., Nekrutenko, A. and Taylor, J. (2010). Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol Chapter 19: Unit 19 10 11-21.
- Feng, J., Liu, T. and Zhang, Y. (2011). Using MACS to identify peaks from ChIP-Seq data. Curr Protoc Bioinformatics Chapter 2: Unit 2 14.
- Feng, J., Liu, T., Qin, B., Zhang, Y. and Liu, X. S. (2012). Identifying ChIP-seq enrichment using MACS. Nat Protoc 7(9): 1728-1740.
- Giardine, B., Riemer, C., Hardison, R. C., Burhans, R., Elnitski, L., Shah, P., Zhang, Y., Blankenberg, D., Albert, I., Taylor, J., Miller, W., Kent, W. J. and Nekrutenko, A. (2005). Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15(10): 1451-1455.
- Lee, K. L., Buckley, H. R. and Campbell, C. C. (1975). An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida Albicans. Sabouraudia 13(2): 148-153.
- Znaidi, S., Nesseir, A., Chauvel, M., Rossignol, T. and d'Enfert, C. (2013). A comprehensive functional portrait of two heat shock factor-type transcriptional regulators involved in Candida albicans morphogenesis and virulence. PLoS Pathog 9(8): e1003519.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Znaidi, S., Proux, C., Weber, S., Drouin, S., Robert, F., Raymond, M., Coppée, J. and d’Enfert, C. (2014). ChIP-Seq in Candida albicans. Bio-protocol 4(12): e1158. DOI: 10.21769/BioProtoc.1158.
- Znaidi, S., Nesseir, A., Chauvel, M., Rossignol, T. and d'Enfert, C. (2013). A comprehensive functional portrait of two heat shock factor-type transcriptional regulators involved in Candida albicans morphogenesis and virulence. PLoS Pathog 9(8): e1003519.
Category
Systems Biology > Genomics > ChIP-seq
Microbiology > Microbial biochemistry > Protein > Interaction
Biochemistry > Protein > Immunodetection > ChIP
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









