- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In organello Protein Synthesis
Published: Vol 4, Iss 12, Jun 20, 2014 DOI: 10.21769/BioProtoc.1157 Views: 14193
Reviewed by: Ru Zhang

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Phosphopeptide Purification Protocol for the Moss Physcomitrella paten
Xiaoqin Wang and Yikun He
Jul 20, 2015 9553 Views

Indirect Immunofluorescence Assay in Chlamydomonas reinhardtii
Takashi Yamano and Hideya Fukuzawa
Jul 5, 2016 13302 Views

Tandem Purification of His6-3x FLAG Tagged Proteins for Mass Spectrometry from Arabidopsis
He Huang and Dmitri Anton Nusinow
Dec 5, 2016 13587 Views
Abstract
In organello protein synthesis method allows the analysis of mitochondrial translation products. The principle of this method relies on incubation of isolated intact mitochondria with radiolabeled amino acids such as 35S methionine. After protein synthesis, the radiolabeled translation products are subsequently separated by SDS polyacrylamide gel electrophoresis and analysed by autoradiography. For in organello analysis of protein synthesis, the isolated intact mitochondria must retain their bioenergetics capacity, and in consequence be fully functional and able to perform coupled respiration. This in turn requires a quick and gentle purification of mitochondria during their isolation.
Keywords: In organello methodMaterials and Reagents
- Youngest leaves harvested from 9- to 10-week-old Arabidopsis thaliana
- Common chemicals
Klorin, NaCl, Tween-20, sucrose, tetrasodiumpyrophosphate, PVP-40, EDTA, KH2PO4, sodium ascorbate, L-cysteine, TES, BSA, GTP, mannitol, KCl, DTT, HEPES, MgCl2, sodium acetate, ADP, malic acid, pyruvate, puromycin, L-methionine, isopropanol, acetic acid, Commassie Blue G-250, glycerol, SDS, Tris, β-mercaptoethanol, bromophenol blue - Sand (50-70 mesh particle size) (Sigma-Aldrich, catalog number: 274739 )
- Percoll (pH 8.5-9.5) (25 °C) (Sigma-Aldrich, catalog number: P1644 )
- L-35S methionine (HARTMANN ANALYTIC, catalog number: SRM-01H )
- Amino acid mixture without L-methionine (Promega Corporation, catalog number: L9961 )
- DC Protein Assay Kit (Bio-Rad Laboratories, catalog number: 500-0112 )
- 12% SDS-polyacrylamide gel (Leammli electrophoresis system)
- 10% chlorox solution (see Recipes)
- Grinding medium (see Recipes)
- 2x wash buffer (see Recipes)
- Synthesis mix (see Recipes)
- Stop solution (see Recipes)
- Isopropanol fixing solution (see Recipes)
- Rapid Coomassie Blue G-250 staining solution (see Recipes)
- Destaining solution (see Recipes)
- 1x solubilzation buffer (see Recipes)
Equipment
- Mortar
- Miracloth (Calbiochem®, catalog number: 475855-1R )
- Polycarbonate centrifuge tubes with round bottom (30 ml and 90 ml)
- 1.5 ml microcentrifuge tubes (SARSTEDT AG, catalog number: 72.690.001 )
- Paint brushes
- Tubes with round bottom (SARSTEDT AG, catalog number: 55.484.001 )
- Pasteur pipette
- Gradient former model 485 (Bio-Rad Laboratories, catalog number: 165-4120 )
- Peristaltic pump –PumpP-1 (GE Healthcare, catalog number: 18-111--91 )
- Centrifuges 1K15 and 3K18 (Sigma-Aldrich)
- Microcentrifuges (Eppendorf, catalog number: 5452 000.018 )
- Incubator shaker (IKA KS4000i control shaker, catalog number: 3510001 )
- Spectrophotometer UV-1800 (Schimadzu, catalog number: 206-25400-32 )
- SDS-PAGE system - Mini-Protean Tetra Cell (Bio-Rad Laboratories, catalog number: 165-8000 )
- Slab Gel dryer SGD5040 (Thermo Fisher Scientific, catalog number: SGD5040-230 )
- Radioactive room
- Carestream Kodak BioMax MR film (Sigma-Aldrich, catalog number: Z353949-50EA )
Procedure
- Isolation of mitochondria from leaves of Arabidopsis by a modification of the method described by Lister et al., (2007).
Note: All subsequent steps perform at 4 °C in sterile conditions.- Prepare 10% chlorox solution and store in cold room.
- Immediately prior to isolation, freshly add the following ingredients to previously prepared (see Recipe 2 below) Grinding medium and 2x wash buffer.The remaining 50 ml of prepared 2x wash buffer (see Recipe 3 below) leave without BSA.
Grinding medium 300 ml 1% (w/v) BSA 3.00 g 18 mM sodium ascorbate 1.06 g 20 mM L-cysteine 0.74 g 2 x wash buffer 150 ml 0.2% (w/v) BSA 0.30 g
Put these buffers in the cold room. - Prepare heavy and light gradient solutions (35 ml). These recipes are sufficient for 4 gradient tubes.
HEAVY (4.4% PVP-40)LIGHT (0% PVP-40)2x wash buffer 17.5 ml Percoll 9.8 ml 20% (w/v) PVP-40) 7.7 ml Put these solutions in the cold room.2x wash buffer 17.5 ml Percoll 9.8 ml MiliQ water 7.7 ml - Immediately after making a heavy and light gradient solutions prepare 1x wash buffer with BSA. For this purpose mix the residual volume of 2x wash buffer with BSA with the same volume of MiliQ water to obtain 1x wash buffer with BSA. Store this buffer in cold room.
- Weigh 25 g youngest leaves harvested from 9- to 10-week-old Arabidopsis thaliana growing under short-day conditions (SD, 22 °C) per sample.
- Sterilize leaves for 5 min in 10% cold chlorox solution. Then rinse the leaves 2-3 times for 1 min in cold sterile MiliQ water. Store leaves on ice, in dark.
Note: Store leaves in dark avoid activation of photosynthesis inside chloroplasts (which can hamper the isolation of purified mitochondria). - Put the leaves in 100 ml Grinding medium and cut the leaves with scissors. Then add 2.5 g of sterile sand (50-70 mesh particle size) and grind leaves energetically in a mortar for 30 sec.
- Filter homogenate through four layers of Miracloth into a conical flask.
- Transfer filtered homogenate (volume of Grinding medium + volume of grinding tissue) to 2 pre-chilled plastic centrifuge tubes (volume of tubes 90 ml).
- Centrifuge for 5 min at 2,450 x g at 4 °C → pellets cell debris and nuclei (general scheme of isolation mitochondria is presented in Figure 1).
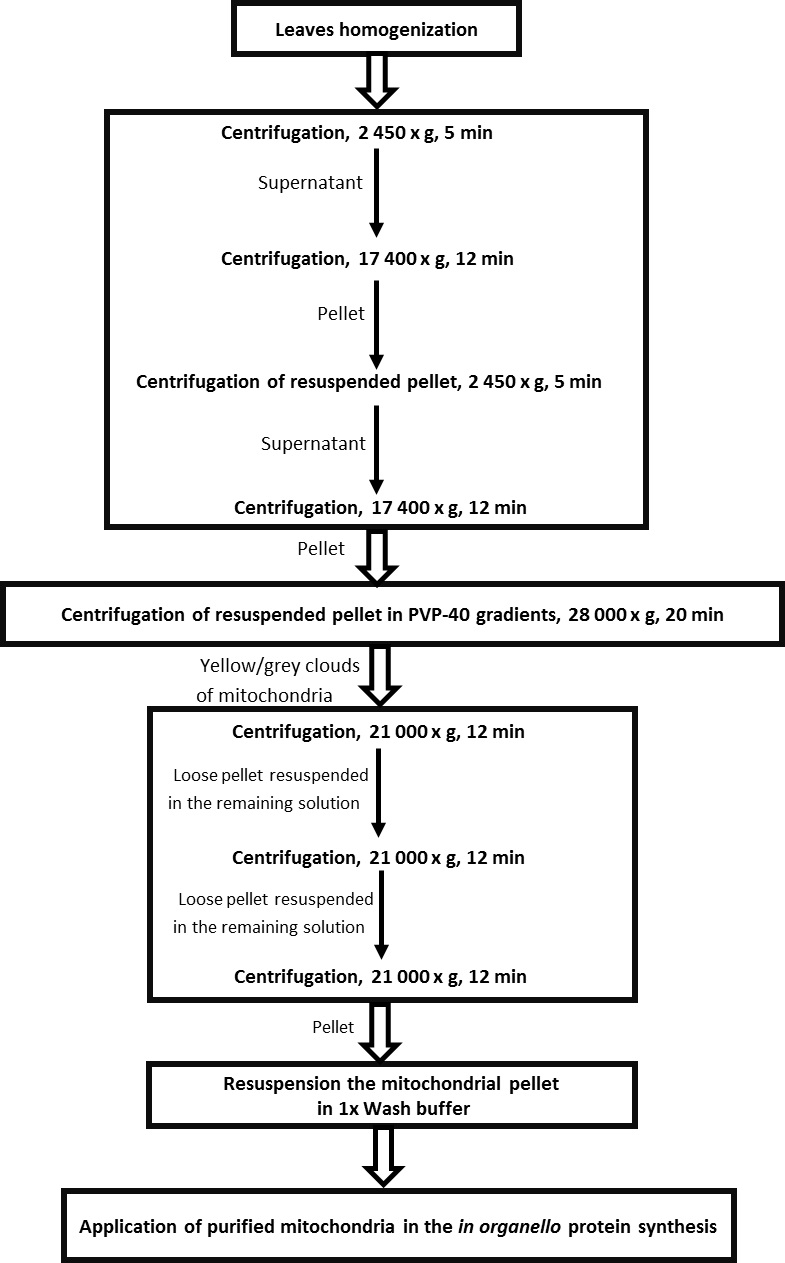
Figure 1. Scheme for the isolation of mitochondria from leaves of Arabidopsis thaliana - Transfer supernatant to new tubes (volume of tubes 90 ml) and centrifuge for 12 min at 17,400 x g at 4 °C → pellets mitochondria, peroxisomes, etc.
- Discard supernatant and resuspend pellets in residual supernatant using a pre-wet with Grinding medium small paintbrush. Gently mix the residual supernatant touching with use of paintbrush the bottom of the tubes (volume of tubes 90 ml) to completely dissolve the pellet of mitochondria.
- Fill the tubes with 1x wash buffer with BSA and repeat centrifuge from steps 10-11 in order to pellet purified mitochondria.
- In the meantime make PVP-40 gradients in pre-chilled centrifuge tubes (volume of tubes 30 ml) using a gradient former model 485 (see Figure 2).
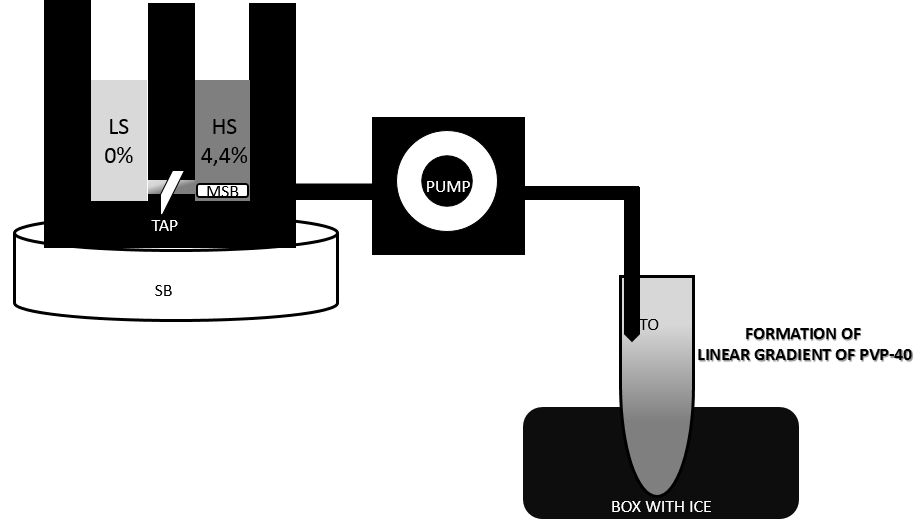
Figure 2. Preparation of linear PVP-40 gradient with a gradient former. Set up tubes on ice and tape outflow tubes (TO) to the inside of the tubes. Close off the connection (TAP) between chambers. Pour 8,75 ml light gradient solution into chamber without tubing outlet. Pour 8,75 ml heavy gradient solution into chamber with tubing outlet and place a small magnetic stir bar (MSB) in this chamber, place gradient former on stirring block (SB). A magnetic stir bar should rotate at a speed sufficient to ensure complete mixing. Set peristaltic pump and allow for a moment only heavy solution to run. Open connection (TAP) between the chambers and allow solution to mix. It is important that the end of the outflow tube (TO) is held below the level of the gradient former so that solution will flow by gravity to down the side of a tubes held below the apparatus. - After centrifugation from step 13 discard supernatant and resuspend pellets in 1.75 ml of 1x wash buffer with BSA using a small paintbrush. Combine the two resuspended pellets of the same type to one tube.
- Load sample carefully on PVP-40 gradient.
- Balance tubes and centrifuge for 20 min at 28,000 x g at 4 °C with the brakes off → separation of mitochondria from chloroplasts and thylakoids.
- After centrifugation mitochondria should form a light yellow/grey clouds at the bottom of the tubes. Carefully remove and discard the layer above the mitochondrial fraction by aspiration with a pasteur pipette.
- Collect mitochondrial fraction with use of truncated tip to a six 1.5 ml Eppendorf tubes filling with 1 ml of 1x wash buffer with BSA. To each tube collect 2 x 200 µl of mitochondrial fractions.
Note: Use of truncated tips allow to collect dense and cloudy mitochondrial fractions. - Centrifuge for 12 min at 21,000 x g at 4 °C.
- Remove supernatant carefully by pipette. Pellet might sit very loose.
- Resuspend gently pellets in all Eppendorf tubes in the remaining medium by snapping the tube. Combine the resuspended pellets of the same type to two arbitrarily chosen tubes.
- Fill these two tubes with 1x wash buffer without BSA and centrifuge for 12 min at 21,000 x g at 4 °C.
- Remove supernatant and resuspend pellets in the remaining medium by gently snapping the tube. Combine the two resuspended pellets of the same type to one tube.
Note: The purpose of this step was purification of isolated mitochondria from Percoll. First the washes in more Eppendorf tubes allowed to discard the remaining contamination from gradients. In the next centrifugation steps the number of Eppendorf tubes were reduced to the one, containing purified mitochondria. - Fill final tube with 1x wash buffer without BSA-centrifuge for 12 min at 21,000 x g at 4 °C.
- Remove supernatant and resuspended mitochondria in 30 µl of 1x wash buffer without BSA.
- Keep mitochondrial fraction on ice to use immediately in the in organello protein synthesis.
- Prepare 10% chlorox solution and store in cold room.
- Protein assay
Determine the mitochondrial protein levels by DC Protein Assay Kit II, follow the instructions provided by the manufacturer. Estimate the protein concentrations for two independent amount of sample by comparison to a standard curve generated by the measurement of BSA of known concentrations. Using standard procedure, the assay is used with samples having protein concentrations between 0.2 and 1.5 mg/ml. The concentrations of mitochondrial proteins which we obtained are between 20-25 µg/µl (mitochondrial fraction dissolved in 30 µl of 1x wash buffer). - In organello protein synthesis
For each mitochondrial preparation, set up two labeling reactions: one for the synthesis mix and one for control synthesis mix, to estimate the bacterial contamination. Use sodium acetate in control reactions instead of malic acid and pyruvate. Sodium acetate is a nonoxidizable substrate that can be utilized by bacteria but not by mitochondria, hence amino acid incorporation in its presence gives an indication of bacterial contamination.- Prepare pre-chilled disposable, sterile 3.5 ml tubes with round bottom .
- Add 25 µl amino acids (without L-methionine) and 1 mg BSA to 1 ml of Synthesis Mix with malic acid and pyruvate or 1 ml of Control Synthesis Mix with sodium acetate. BSA protects the freshly isolated mitochondria. Keep on ice.
- Pipette to the pre-chilled tubes 100 µl of Synthesis Mix or 100 µl Control Synthesis Mix.
- Add to each tubes 150 µg of freshly isolated mitochondrial fractions.
- Add 30 µCi 35S L-methionine -1.5 µl (20 mCi/ml) per sample and incubate at 25 °C for 30 or 60 min in incubator shaker (speed 180 rpm).
Note: During the assay keep the mitochondria well oxygenated using a shaker, to avoid the risk of anoxia due to their sedimentation to the bottom of the tube. In our experience, increasing the incubation times beyond 60 min does not increase the labeling of mitochondrial products. - Following incubation reactions are stopped by addition 350 µl of ice cold 1x Wash buffer without BSA containing 10 mM unlabelled L-methionine and puromycin (50 µg/ml).
- Mix using pipette and transfer sample to new 1.5 ml Eppendorf tubes. Keep on ice.
- Pellet the mitochondrial fractions at 21,000 x g for 5 min in centrifuge at 4 °C.
- Remove the supernatant (containing the free 35S-methionine) and resuspend the mitochondrial pellet in 30 µl of 1x wash buffer without BSA.
- Mitochondria may be frozen on dry ice and stored at -80 °C or resuspended in 20-30 µl of electrophoresis sample buffer for immediate SDS-PAGE analysis (see D).
- Prepare pre-chilled disposable, sterile 3.5 ml tubes with round bottom .
- SDS-Polyacrylamide gel electrophoresis and autoradiography
- Solubilize entire mitochondrial sample in a 1x solubilzation buffer by heating for 5 min at 95 °C.
- Analyse the samples on 12% SDS-polyacrylamide gel (Leammli electrophoresis system).
- After electrophoresis gently agitate gel on a platform shaker in isopropanol fixing solution for 15 min.
- Discard fixing solution and gently agitate gel in Rapid Commasie Blue G-250 staining solution for 30 min.
- Remove staining solution and gently agitate gel in destaining solution for 40-60 min until a clear background with blue protein bands appears.
- Agitate gel in water 3 times for 5 min.
- After rinsing agitate gel in water containing 5% (v/v) of glycerol for 7 min. Agitation the gel in water containing glycerol may protect gel before cracking during drying.
- Dry gel onto a Whatman 3 MM paper using gel dryer, and expose for minimum 3 days to a Kodak BioMax MR film.
- Solubilize entire mitochondrial sample in a 1x solubilzation buffer by heating for 5 min at 95 °C.
Representative data
- The example of autoradiogram of proteins synthesized in organello in Arabidopsis mitochondria is presented in below figure (Figure 3).
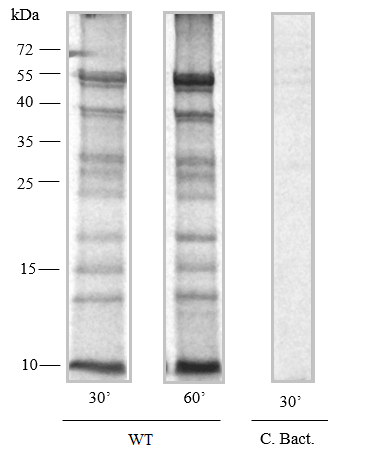
Figure 3. In organello protein synthesis. Fragments of autoradiogram of proteins synthesized in organello for 30 and 60 min by wild-type mitochondria isolated from Arabidopsis (Kwasniak et al., 2013). After protein synthesis, 25 µg of mitochondria were fractionated on 12% (w/v) SDS-PAGE. The lane marked C. Bact. is a specific control performed in the presence of a sodium acetate- substrate for bacterial translation.
Recipes
Note: All the buffers and solutions are prepared with double-distilled water and sterilized by autoclave.
- 10% chlorox solution (1 L)
Klorin 100 ml NaCl 10 g Tween-20 15 drops - Grinding medium (300 ml)Dissolve well in ~250 ml of MiliQ water, adjust pH to 7.5 with HCl and then make up to 300 ml with MiliQ water.
0.3 M sucrose 30.80 g 25 mM tetrasodiumpyrophosphate (anhydrous) 1.99 g 2 mM EDTA (disodium salt) 0.22 g 10 mM KH2PO4 0.40 g 1% (w/v) PVP-40 3.00 g - 2x wash buffer (200 ml)
Dissolve well in ~150 ml of MiliQ water, adjust pH to 7.5 with NaOH and then make up to 200 ml with MiliQ water.0.6 M sucrose 41.0 g 20 mM TES 0.9 g - Synthesis mix (20 ml) Mix solution and divide to
5 mM KH2PO4 13.61 mg 2 mM GTPNa 20.93 mg 0.4 M mannitol 1457.00 mg 60 mM KCl 89.46 mg 2 mM DTT 6.17 mg 50 mM HEPES 238.31 mg 10 mM MgCl2 40.66 mg 4 mM ADP (K) 40.10 mg pH 7.0 - 15 ml synthesis mix → addStored at -80 °C
10 mM malic acid 26.71 mg 1 mM pyruvate (Na) 1.65 mg Filter sterilize (0.22 µm pore size) 15 x 1 ml - 5 ml control synthesis mix → add
20 mM Na-sodium acetate 8.20 mg Filter sterilize (0.22 µm pore size) 5 x 1 ml Stored at -80 °C
- 15 ml synthesis mix → add
- Stop solution (50 ml)
10 mM L-methionine (0.0746 g) dissolved in 50 ml of 1x wash buffer without BSA - Isopropanol fixing solution (1 L)
Isopropanol 250 ml Acetic acid 100 ml Water 650 ml - Rapid Coomassie Blue G-250 staining solution (1 L)
Acetic acid 100 ml Water 900 ml Commassie brilliant blue G-250 60 mg - Destaining solution (1 L)
Acetic acid 100 ml Water 900 ml - 1x solubilzation buffer
2% (v/v) sodium dodecyl sulfate (SDS)
10% glycerol
62.5 mM Tris-HCl (pH 6.8)
0.002 % (w/v) bromophenol blue
5% β-mercaptoethanol
Note: Indicated concentrations of components of solubilization buffer are final concentrations.
Acknowledgments
Functional mitochondria for in organello protein synthesis were isolated from leaves of Arabidopsis by a modification of the methods described by Lister et al., 2007 and Millar et al., 2007. In organello protein synthesis was performed for the first time on mitochondria isolated from leaves of Arabidopsis thaliana as described in the paper Kwasniak et al., 2013, and was based on previously published papers including Boutry et al., 1984. We are grateful to Christopher J. Leaver for several highly relevant papers that allow us to know details about the background of the in organello protein synthesis procedure.
References
- Boutry, M., Faber, A. M., Charbonnier, M. and Briquet, M. (1984). Microanalysis of plant mitochondrial protein synthesis products: Detection of variant polypeptides associated with cytoplasmic male sterility. Plant Mol Biol 3(6): 445-452.
- Echan, L. A. and Speicher, D. W. (2002). Protein detection in gels using fixation. Curr Protoc Protein Sci Chapter 10: Unit 10 15.
- Fernandez-Silva, P., Acin-Perez, R., Fernandez-Vizarra, E., Perez-Martos, A. and Enriquez, J. A. (2007). In vivo and in organello analyses of mitochondrial translation. Methods Cell Biol 80: 571-588.
- Grohmann, L. (1995). In organello protein synthesis. Methods Mol Biol 49: 391-397.
- Hakansson, G., van der Mark, F., Bonnett, H. T. and Glimelius, K. (1988). Variant mitochondrial protein and DNA patterns associated with cytoplasmic male-sterile lines of Nicotiana. Theor Appl Genet 76(3): 431-437.
- Kwasniak, M., Majewski, P., Skibior, R., Adamowicz, A., Czarna, M., Sliwinska, E. and Janska, H. (2013). Silencing of the nuclear RPS10 gene encoding mitochondrial ribosomal protein alters translation in arabidopsis mitochondria. Plant Cell 25(5): 1855-1867.
- Lister, R., Carrie, C., Duncan, O., Ho, L. H., Howell, K. A., Murcha, M. W. and Whelan, J. (2007). Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell 19(11): 3739-3759.
- Millar, A. H., Liddell, A. and Leaver, C. J. (2007). Isolation and subfractionation of mitochondria from plants. Methods Cell Biol 80: 65-90.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kwasniak-Owczarek, M. and Janska, H. (2014). In organello Protein Synthesis. Bio-protocol 4(12): e1157. DOI: 10.21769/BioProtoc.1157.
Category
Plant Science > Plant biochemistry > Protein > Labeling
Cell Biology > Organelle isolation > Mitochondria
Molecular Biology > Protein > Detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link