- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
MTV1 Pull-down Assay in Arabidopsis
Published: Vol 4, Iss 12, Jun 20, 2014 DOI: 10.21769/BioProtoc.1152 Views: 13159
Reviewed by: Ru Zhang

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Streamlining Protein Fractional Synthesis Rates Using SP3 Beads and Stable Isotope Mass Spectrometry: A Case Study on the Plant Ribosome
Dione Gentry-Torfer [...] Federico Martinez-Seidel
May 5, 2024 2870 Views

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1989 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1772 Views
Abstract
This protocol is an example of how to analyse suspected interactions between proteins using a pull-down assay (Sauer et al., 2013). A bait protein of interest (in this case, MTV1 of Arabidopsis thaliana) is fused to a GST tag and expressed in bacteria. The protein is isolated and allowed to bind to a matrix of glutathione-conjugated agarose beads via the GST-tag. Unspecifically binding proteins from the bacterial lysate are removed from the matrix. A native plant protein extract is then passed over the matrix and binding between the bait GST-MTV1 and prey proteins can occur. Extensive washes remove unspecifically bound proteins and finally, bait and prey proteins are released from the beads. Immunoblot analysis is then used to identify the proteins that bound to GST-MTV1. Importantly, a negative control consisting of the GST-tag alone is analysed in parallel to exclude the possibility that prey protein binding to the GST-MTV1 bait was due to the GST-tag.
Keywords: Pull-downMaterials and Reagents
- BL21 Escherichia coli (E. coli) cells containing a plasmid for expression of the recombinant GST-MTV1 fusion protein (the bait)
Note: In this exemplary case, the MTV1 coding sequence was cloned into a modified pGEX-2T plasmid (General Electric Company, catalog number: 28-9546-53 ), in which the multiple cloning site had been replaced by a Gateway cloning cassette (Note 1).
- BL21 E. coli cells expressing the GST tag alone as negative control
- Murashige and Skoog medium mix with vitamins and MES buffer (Duchefa Biochemie BV, catalog number: M0255.0010 )
- Glutathione agarose (Sigma-Aldrich, catalog number: G4510 )
- Complete inhibitor (EDTA free) (Roche Diagnostics, catalog number: 11 873 580 001 )
- Phenylmethylsulfonyl fluoride PMSF (e.g. Sigma-Aldrich, catalog number: P7626 )
- Triton X-100 (Sigma-Aldrich, catalog number: T8787 )
- Carbenicillin (e.g. Sigma-Aldrich, catalog number: C9231 ) (Note 2)
- Isopropyl β-D-1-thiogalactopyranoside (IPTG) (e.g. Sigma-Aldrich, catalog number: I6758 )
Note: Should be prepared as aqueous stock solution of 1 M and stored at -20 °C.
- Sodium dodecyl sulfate (SDS) (e.g. Sigma-Aldrich, catalog number: L3771 , or any other supplier)
Note: An aqueous stock solution of 20% (weight/volume) can be prepared and sterilized by autoclaving at 121 °C for 15 min.
- Glycerol (e.g. Sigma-Aldrich, catalog number: G5516 , or any other supplier)
- Tris (hydroxymethyl) aminomethane (Tris) (e.g. Sigma-Aldrich, catalog number: 252859 , or any other supplier)
- Yeast extract (e.g. Sigma-Aldrich, catalog number: Y1625 , or any other supplier)
- Tryptone (e.g. Sigma-Aldrich, catalog number: 95039 , or any other supplier)
- Anti-GST polyclonal antibody (optional) (Carl Roth, catalog number: 3998 )
- Anti-CHC monoclonal antibody (optional) (BD Biosciences, catalog number: 610499 )
- Liquid nitrogen
- NaCl (any supplier)
- KCl (any supplier)
- Na2HPO4 (any supplier)
- KH2PO4 (any supplier)
- β-mercaptoethanol
- Liquid grown Arabidopsis seedlings of 6-8 days (see Recipes)
- MS medium (see Recipes)
- PBS (see Recipes)
- Wash buffer (see Recipes)
- Extraction buffer (see Recipes)
- Sample loading buffer (see Recipes)
- Liquid Lysogeny Broth (LB) growth medium (see Recipes)
Equipment
- Microcentrifuge for 1.5 and 2 ml standard reaction tubes (any manufacturer)
Note: Either refrigerated or situated in a 4 °C cold-room, should be able to reach 16,000 x g.
- Refrigerated centrifuge for 50 ml conical “Falcon” type tubes (any manufacturer)
Note: Should reach 4 °C and 3,000 x g.
- Sonicator device (micro tip sonotrode type)
Note: We use the labsonic model of B. Braun, which is, however, no longer produced. But any tip style sonicator device that is suitable for small volumes (2-5 ml) will work, for example the UP100H device coupled to the MS3 sonotrode (Hielscher Ultrasound Technology).
- Poly-Prep Chromatography columns (Bio-Rad Laboratories, catalog number: 731-1550 )
- Erlenmeyer flasks
- Paper towels
- 1.5 ml microcentrifuge tubes capable of supporting 16,000 x g (any supplier)
- 50 ml polypropylene conical centrifuge tubes “Falcon” type (any supplier)
- 0.20 µm filter unit (e.g. Minisart®, Sartorious, catalog number: 17597 ) plus compatible 5 ml syringe
- Shaking incubator for bacteria 37 °C (any manufacturer)
- Shaking incubator 25 °C or orbital shaker at room temperature (any manufacturer)
- End-over-end (orbital) mixer (any manufacturer)
- Spectrophotometer capable of measuring optical density at 600 nm (any manufacturer)
- Mortar and pestle (about 10 cm diameter) (any manufacturer)
Procedure
- Planning ahead
The pellets of bacterial culture expressing GST-MTV1 and GST can be prepared any time in advance, as this material can be stored at -80 °C for several weeks. The plant extract is preferentially prepared on the day of the actual pulldown experiment. To generate this plant material, calculate a total of 8-10 days (from seed sterilization to harvest). The pulldown experiment can be carried out in one day.
- Protein expression in bacteria
- From single bacterial colonies (or verified glycerol stocks) of GST-MTV1 (bait) and GST (negative control) expressing bacteria, grow overnight cultures in 10 ml LB with appropriate antibiotic selection (in this case, 100 μg/ml carbenicillin). Use 50 ml Falcon type tubes and grow cultures in a shaking incubator at 37 °C with agitation of at least 200 rpm. Tubes should not be fully closed to allow gas exchange.
- Next day, prepare two Erlenmeyer flasks (500 ml volume) each with 150 ml LB + 100 μg/ml carbenicilin and add the MTV1-GST and GST pre-culture, respectively. Grow at 25 °C on an orbital shaker at least at 200 rpm, until OD600 reaches 0.6. Growing at 25 °C helps to produce the proteins in soluble form.
- To induce the expression of GST-MTV1 and GST, add IPTG to a final concentration of 1 mM and continue to grow the cultures at 25 °C for another 3-4 h.
- Distribute each culture into three 50 ml tubes and centrifuge them at 4 °C at 3,000 x g for 5 min.
- Discard the supernatant and gently resuspend the pellet in ice cold PBS, then centrifuge again as above.
- Discard the supernatant and store the pellets at -80 °C. Pellets can be stored for several weeks. For one pulldown experiment, only one of the pellets (corresponding to 50 ml of bacterial culture) is used.
- From single bacterial colonies (or verified glycerol stocks) of GST-MTV1 (bait) and GST (negative control) expressing bacteria, grow overnight cultures in 10 ml LB with appropriate antibiotic selection (in this case, 100 μg/ml carbenicillin). Use 50 ml Falcon type tubes and grow cultures in a shaking incubator at 37 °C with agitation of at least 200 rpm. Tubes should not be fully closed to allow gas exchange.
- Extract protein from bacteria
- Take one pellet of GST-MTV1 and GST (corresponding to 50 ml bacterial culture) and resuspend each in 2.5 ml ice cold extraction buffer.
- Use a sonicator to disrupt the bacterial cells until they are lysed, which is indicated by reduced opacity and increased viscosity. This has to be carried out on ice, and it is recommended to sonicate in intervals (e.g. 10 sec sonicating followed by 10 sec on ice and so on). Care has to be taken not to sonicate with too high intensity. If foaming or the formation of a white precipitate is detected, the intensity needs to be lowered.
- Transfer the lysed bacteria solution to 1.5 ml microcentrifuge tubes and centrifuge at 4 °C, 16,000 x g for 20 min.
- Take one pellet of GST-MTV1 and GST (corresponding to 50 ml bacterial culture) and resuspend each in 2.5 ml ice cold extraction buffer.
- Reconstitute glutathione beads
- In the meantime, reconstitute the glutathione agarose beads. For each GST-MTV1 and GST, weigh 10 mg of the dry beads and add 2 ml washing buffer. Let the beads swell for 10 min while vortexing occasionally.
- Load the slurries onto two Poly-Prep columns, place them upright in an appropriate holder and let the buffer drain through the bottom opening by gravity.
Note: Do not centrifuge or apply vacuum to increase flow rate! This applies to all subsequent washing steps.
- Then wash the column with 10 ml washing buffer, again letting the column drain by gravity.
- In the meantime, reconstitute the glutathione agarose beads. For each GST-MTV1 and GST, weigh 10 mg of the dry beads and add 2 ml washing buffer. Let the beads swell for 10 min while vortexing occasionally.
- Protein binding to glutathione beads
Of the centrifuged bacterial lysates, take 150 µl of the supernatant and mix with 75 µl sample loading buffer, boil for 5 min and store at -20 °C, this will serve later as a control to check protein expression and integrity after bacterial lysis, for instance by immunoblot analysis using anti-GST antibody. The rest of the supernatant is loaded onto the columns, one for GST-MTV1 and one for GST. If viscosity of the lysate seems high, 1-2 ml of extraction buffer can be added. Close column in- and outlets and place them onto a rotating wheel at 4 °C for 2 h. Let the lysate drain by gravity, then wash the columns 3 times with 10 ml washing buffer at 4 °C.
- Plant extract
- Blot the liquid grown Arabidopsis seedlings dry, using paper towels. Weigh 0.7 g and transfer to a mortar.
- Freeze with liquid nitrogen and grind to a fine powder with a pestle.
- Add 4 ml extraction buffer and let slowly thaw, grinding the sample even more.
- Transfer the sample to 1.5 ml microfuge tubes and centrifuge for 20 min at 4 °C at 16,000 x g.
- Pass supernatant through a 0.20 µm filter syringe and keep extract on ice. Per column, 1 ml extract will be needed.
- Blot the liquid grown Arabidopsis seedlings dry, using paper towels. Weigh 0.7 g and transfer to a mortar.
- Pulldown
- Close column outlets.
- Per column, mix 1 ml extract with 1 ml extraction buffer and add it onto the washed column.
- Close column inlets and place them onto a rotating wheel at 4 °C for 1.5 h.
- Then open the column in- and outlets and collect the flowthrough.
- Mix 150 µl of the flowthrough of each column with 75 µl sample loading buffer and boil for 5 min, keep at -20 °C. These samples are the flowthrough, containing all the unbound plant proteins (and usually some bacterial proteins as well).
- Wash the columns 3 times with 10 ml washing buffer at 4 °C, again letting drain by gravity.
- The bead volume is now about 100 µl. Close the column outlet and re-suspend the beads in 200 µl washing buffer.
- Take 150 µl of the bead slurry and add 75 µl sample loading buffer. Boil the sample for 5 min, mix vigorously and centrifuge for 5 min at 16,000 x g.
- Pass the supernatant to a fresh tube and store at -20 °C. These samples are the eluates which contain the proteins that bound to GST-MTV1 or GST, respectively, plus the bait proteins themselves.
- Typically, the flowthrough and the eluate samples are loaded next to each other on a immunoblot for both GST and GST-MTV1 (see Figure 1). It is often advisable to correct for different protein amounts in flowthrough and eluate, a good starting point is to load 5 µl flowthrough and 20 µl eluate, but this has to be determined empirically.
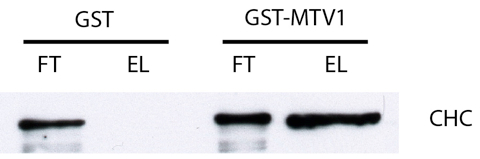
Figure 1. Typical result of an immunoblot usig anti-clathrin heavy chain (CHC) antibody (approx. 190 kDA). In the flowthrough (FT) of both GST and GST-MTV1 the protein is detected, indicating that CHC was present at equal levels during the incubation. After washing, only the eluate (EL) of GST-MTV1 produces a signal, indicating that CHC bound specifically to MTV1. In this blot, 5 µl of FT sample and 20 µl of EL sample were loaded to compensate for different protein levels.
- A protein which binds specifically to GST-MTV1 will be detected in both flowthrough samples, but only in the eluate sample of GST-MTV1. This is for example the case for clathrin heavy chain (CHC), which can be detected using a commercial anti-CHC antibody (see Figure 1).
- A protein which is detected in both eluate samples is binding unspecifically to either the GST tag or the glutathione-agarose matrix. In this case, it can help to increase duration and number of washing steps or use a different washing buffer with higher amounts of salts or detergents from step G 6 onwards.
- Close column outlets.
Notes
- This protocol can in theory be used for any kind of protein fused to the GST tag. However, the conditions likely need to be optimized for each particular case. MTV1 is an example of a soluble protein that is easily expressed in E. coli and does not require any eukaryotic posttranslational modifications, such as glycosylation. Other proteins might require a different expression and buffer system.
- PMSF is highly toxic and very unstable in aqueous solutions, so the preparation has to be done with care. A common practice is to prepare a 200 mM stock solution in methanol, which can be stored at -20 °C for at least half a year.
- The considerably more expensive carbenicillin can be substituted with ampicillin using the same concentration. However, ampicillin is less stable, so carbenicillin is the preferred choice.
- Complete inhibitor comes in the form of small tablets, each good for a volume of 50 ml final buffer. If a smaller volume is desired, the tablet can be dissolved in 1 ml water to produce a 50x stock solution and any surplus can be stored at -20 °C for a couple of weeks without dramatic loss of activity.
Recipes
- Liquid grown Arabidopsis seedlings
- 200-300 seeds of Arabidopsis thaliana (We used Col-0 accession, but can be any accession, mutant or transgenic line.) are sterilized in a 1.5 ml microcentrifuge tube by a 15 min incubation in 70% ethanol with occasional vortexing.
- In a cleanbench, wash the seeds 5 times with 1 ml sterile water. After the last wash, keep the seeds in water, close the tube and keep it in darkness at 4 °C for 2-3 days for stratification.
- The stratified seeds are then passed to a 250 ml Erlenmeyer flask containing 100 ml MS medium and incubated in a plant growth chamber with slight agitation (such as an orbital shaker at 50 rpm). We use a 16 h light/8 h dark regime and 24 °C. Plants are grown for 6-8 days.
- 200-300 seeds of Arabidopsis thaliana (We used Col-0 accession, but can be any accession, mutant or transgenic line.) are sterilized in a 1.5 ml microcentrifuge tube by a 15 min incubation in 70% ethanol with occasional vortexing.
- MS medium (1 L)
2.45 g Murashige and Skoog medium mix with vitamins and MES buffer
10 g sucrose
Add mili-Q water to 1 L
Adjust pH to 5.8 using KOH
- PBS (1 L)
8 g NaCl
0.2 g KCl
1.44 g Na2HPO4
0.24 g KH2PO4
Dissolve in 800 ml mili-Q water
Adjust pH to 7.4 using HCl
Then fill up to 1,000 ml
- Wash buffer
PBS
0.5% Triton X-100
0.5 mM PMSF (made from a stock solution of 200 mM PMSF in methanol ) (Note 3)
- Extraction buffer
Like wash buffer, but with the addition of 1x complete inhibitor (Note 4)
- Sample loading buffer (50 ml)
15 ml 20% SDS aqueous stock solution
15 ml glycerol
15 ml Tris 0.5 M solution (pH 6.8)
1.25 ml mili-Q water
A small amount (spatula tip) of bromphenol blue, just enough to give a medium blue color.
Prior to use, add 75 μl β-mercaptoethanol per 925 μl buffer.
- Liquid Lysogeny Broth (LB) growth medium (1 L)
10 g tryptone
5 g yeast extract
10 g NaCl
Fill to 1 L with mili-Q water
Autoclave at 121 °C for 15 min
Acknowledgments
This protocol was adapted from the previously published paper Sauer et al., 2013. Part of the work was funded by a Ramon y Cajal research stipend to M.S.
References
- Sauer, M., Delgadillo, M. O., Zouhar, J., Reynolds, G. D., Pennington, J. G., Jiang, L., Liljegren, S. J., Stierhof, Y. D., De Jaeger, G., Otegui, M. S., Bednarek, S. Y. and Rojo, E. (2013). MTV1 and MTV4 encode plant-specific ENTH and ARF GAP proteins that mediate clathrin-dependent trafficking of vacuolar cargo from the trans-Golgi network. Plant Cell 25(6): 2217-2235.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sauer, M. (2014). MTV1 Pull-down Assay in Arabidopsis. Bio-protocol 4(12): e1152. DOI: 10.21769/BioProtoc.1152.
Category
Plant Science > Plant biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Expression
Biochemistry > Protein > Interaction > Protein-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










