- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of Pseudokinase-ligand Interaction by a Fluorescence-based Thermal Shift Assay
Published: Vol 4, Iss 11, Jun 5, 2014 DOI: 10.21769/BioProtoc.1135 Views: 13451
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Analytical Gel Filtration for Probing Heavy Metal Transfer between Proteins
Steffen Lorenz Drees and Mathias Lübben
Aug 5, 2016 11109 Views

Hydrogen Deuterium Exchange Mass Spectrometry of Oxygen Sensitive Proteins
Luke Berry [...] Brian Bothner
Mar 20, 2018 10258 Views

Binding Affinity Quantifications of the Bacteriophage Mu DNA Modification Protein Mom Using Microscale Thermophoresis (MST)
Shubha Udupa [...] Shweta Karambelkar
Jul 20, 2022 3706 Views
Abstract
This protocol describes a robust technique for the measurement of pseudokinase-ligand interaction by a fluorescence-based Thermal Shift Assay (TSA). Pseudokinases are kinase-like proteins that have recently emerged as crucial regulators of signal transduction and may therefore represent a novel class of drug targets. Unlike kinases, the catalytic efficiency of pseudokinases is rather poor or non existent, making it difficult to dissect the function of their nucleotide binding sites. Thermal denaturation-based methods have proven to be a powerful method for determining ligand binding capacity to purified pseudokinases and can inform on the potential drugability of the nucleotide binding site. This assay takes advantage of a change in flurorescence arising when a flurorescence dye, in this instance SYPRO® Orange, binds to hydrohobic patches that become exposed when a protein undergoes thermal denaturation. Ligand binding to a protein is known to increase its thermal stability which is reflected by a shift observed in the thermal denaturation curve between the unliganded protein and the liganded protein. This generalized protocol can also be tailored to other protein families. In addition, thermal denaturation-based methods can be used to identify optimal buffer conditions that may increase protein stability.
Materials and Reagents
- Purified protein (stock solution preferably at a concentration above 20 μM) (Murphy et al., 2013)
- 1 M Dithiothreitol (DTT) stock (Astral Scientific, catalog number: C-1029 )
- DMSO (high grade) (Sigma-Aldrich, catalog number: D-1435 )
- MilliQ water
- Nucleotide solutions (10 mM stock prepared in 20 mM Tris) (pH 8)
- Divalent cations salt solutions (50 mM stock in MilliQ water)
- Kinase inhibitor solutions (2 mM stock in 100% DMSO) (such as the pan-kinase inhibitors, Staurosporine, Sigma-Aldrich, catalog number: S5921 )
- Thermal Shift Assay buffer (see Recipes)
- SYPRO® Orange (Sigma-Aldrich, catalog number: S5692 ) (see Recipes)
Equipment
- RT-PCR tubes GST-RG01 (Gene Targets Solutions)
- 1.5 ml microfuge tube
- Qiagen/Corbett Rotor-Gene® 3000 RT-PCR machine (QIAGEN) (Murphy et al., 2013)
Software
- Microsoft Excel or Prism
Procedure
- Thermal Shift Assay (TSA) test run for the determination of optimal amount of protein
- Prepare a dilution series of your protein sample in buffer ranging from 1 to 10 μM final concentration in a total volume of 25 μl. Add 1 μl of 1x SYPRO® Orange to each tube.
- Perform a thermal cycler run using the parameters as described in step 3. Analyze the melt-curve (see step 4) and determine the optimal amount of protein that gives at least 30 fluorescence unit. Reduce amount of protein if the fluorescence signal is saturated.
Notes: Typical final protein concentration is around 5 μM. The Rotor-Gene® 3000 is equiped with a gain optimization function that can be adjusted for optimal signal (refer to manufacturer’s manual).
- Prepare a dilution series of your protein sample in buffer ranging from 1 to 10 μM final concentration in a total volume of 25 μl. Add 1 μl of 1x SYPRO® Orange to each tube.
- Assay
- Start by defining the number of conditions that you need to test, including appropriate controls such as protein sample in buffer only if testing nucleotide binding, or in presence of DMSO if testing compounds. This will determine the total volume of master mix of protein needed to conduct the entire experiment. Each reaction is conducted in a total volume of 25 μl.
- Prepare on ice just prior use a master mix containing your protein in the Thermal Shift Assay buffer using the concentration of protein that gave the peak fluorescence (without signal saturation) in step 1.
- Dispense 23 μl of protein/buffer, add 1 μl of ligand (10 to 200 μM) or 1 μl of DMSO/buffer for the control experiment and 1 μl of diluted SYPRO® Orange (1:100). Prepare tubes in duplicate. If the protein to test is highly unstable at room temperature, it is recommended to prepare the tubes on ice. Nucleotide solutions are typically used at a final concentration of 200 μM final, divalent cations at 1 mM final and kinase inhibitors at 40 μM final.
- Start by defining the number of conditions that you need to test, including appropriate controls such as protein sample in buffer only if testing nucleotide binding, or in presence of DMSO if testing compounds. This will determine the total volume of master mix of protein needed to conduct the entire experiment. Each reaction is conducted in a total volume of 25 μl.
- Thermal cycler program
- Fluorescence-based Thermal Shift Assay can be performed using instruments that combine both sample temperature control and dye fluorescence detection. In this instance, we used the Qiagen/Corbett Rotor-Gene® 3000 RT-PCR machine.
- Start at 25 °C, hold at 25 °C for 2 min, increase temperature of 1 °C/min for 65 cycles (25 °C to 90 °C) reading fluorescence intensity every °C. Return to 25 °C. Excitation is at 470 nm (green chanel) and emission is at 555 nm (yellow chanel).
- Fluorescence-based Thermal Shift Assay can be performed using instruments that combine both sample temperature control and dye fluorescence detection. In this instance, we used the Qiagen/Corbett Rotor-Gene® 3000 RT-PCR machine.
- Data analysis
- Save data as fluorescence intensity vs temperature. Export file in a format suitable for import into Microsoft Excel or Prism for analyses.
- The highest fluorescence density is used as a cutoff for the data, therefore remove all data after the high fluroescence density peak. Fit fluorescence intensity curve to a Boltzmann sigmoidal curve using Prism.
- Obtain the melting temperature (Tm) of the protein in buffer/DMSO, which correspond to the midpoint for the protein unfolding curve. Similarly, obtain the Tm of the protein when ligand is added.
- Calculate ΔTm = Tm ligand - Tm buffer/DMSO. A positive ΔTm indicates that the ligand stabilizes the protein from denaturation, hence binds to the protein. A value ≥ to 2-3 °C is an indicator of ligand binding.
- Below is an example of a typical TSA experiment.
Notes: If PRISM software is not available, non-linear regression analysis of experimental data can be conducted using a Microsoft Excel spreadsheet (Brown, 2001).
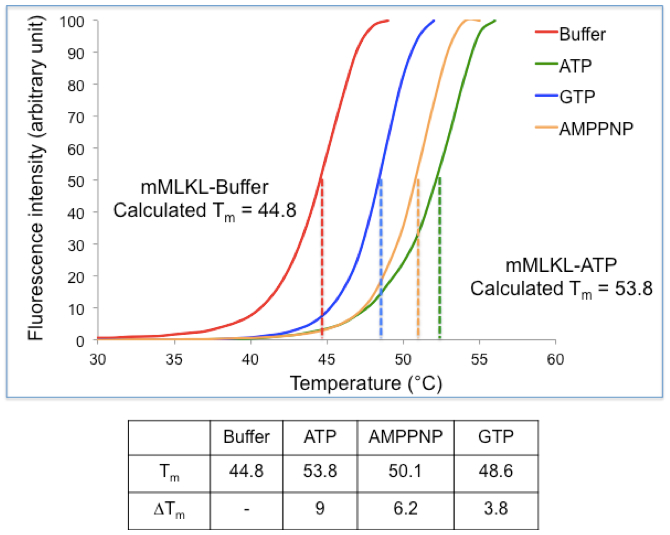
Figure 1. Practical example of a TSA experiment. (Top) Thermal Shift Assay data were obtained for mouse MLKL in the presence and absence of nucleotides. The midpoint for the unfolding transition in the absence of ligand was observed to be 44.8 °C while the Tm shifted to 53.8 °C in presence of ATP, inducing a shift of 9 °C. (Bottom) Table summarizing the Tm and ΔTm values obtained for mMLKL in presence of ATP, GTP and AMPPNP.
- Save data as fluorescence intensity vs temperature. Export file in a format suitable for import into Microsoft Excel or Prism for analyses.
Recipes
- Thermal Shift Assay buffer
The composition of the buffer used to dilute the protein can be optimised according to each protein. The following recipe was used to study pseudokinases.
20 mM Tris (pH 8)
150 mM NaCl
1 mM DTT
- SYPRO® Orange
1 to 100 dilution in 100% high grade DMSO
Notes
- While the thermal denaturation of a single domain proteins mostly results in a single transition phase, multidomain proteins may exhibit distinct transition states. Nevertheless if a ligand binds tightly to one domain, it could result in a change in the thermal denaturation profile, providing a tractable method for assaying ligand binding. There are also cases in which a ligand may bind tightly but does not induce a change in the Tm.
- The protocol described has been adapted to the Qiagen/Corbett Rotor-Gene® 3000 RT-PCR machine. The amount of protein required per experiment and the SYPRO® Orange dilution may need to be adapted depending on the RT-PCR machine used.
Acknowledgments
We thank the Monash University Protein Production Unit for access to the Corbett RT-PCR instrument used for development of the thermal-shift assay. This work was supported by National Health and Medical Research Council (NHMRC) grants (1016647, 461221, 1016701, 637342, 1025594, 1046984) and fellowships to J.M.H., N.A.N. and J.S.; Australian Research Council fellowships to P.E.C., J.J.B. and J.M.M.; and additional support from the Australian Cancer Research Fund, Victorian State Government Operational Infrastructure Support, and NHMRC IRIISS grant (361646). This assay was developed over the course of completing the following studies: Murphy et al. (2013) and Murphy et al. (2014).
References
- Brown, A. M. (2001). A step-by-step guide to non-linear regression analysis of experimental data using a Microsoft Excel spreadsheet. Comput Methods Programs Biomed 65(3): 191-200.
- James, M. M., Isabelle, S. L., Joanne, M. H., Maria, C. T., Samuel, N. Y., Pooja, S., Guillaume, L., Warren, S. A., Jeffrey, J. B. and John, S. (2014). Insights into the evolution of divergent nucleotide-binding mechanisms among pseudokinases revealed by crystal structures of human and mouse MLKL. Biochem J 457(3): 369-377.
- Murphy, J. M., Czabotar, P. E., Hildebrand, J. M., Lucet, I. S., Zhang, J. G., Alvarez-Diaz, S., Lewis, R., Lalaoui, N., Metcalf, D., Webb, A. I., Young, S. N., Varghese, L. N., Tannahill, G. M., Hatchell, E. C., Majewski, I. J., Okamoto, T., Dobson, R. C., Hilton, D. J., Babon, J. J., Nicola, N. A., Strasser, A., Silke, J. and Alexander, W. S. (2013). The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 39(3): 443-453.
- Murphy, J. M., Zhang, Q., Young, S. N., Reese, M. L., Bailey, F. P., Eyers, P. A., Ungureanu, D., Hammaren, H., Silvennoinen, O., Varghese, L. N., Chen, K., Tripaydonis, A., Jura, N., Fukuda, K., Qin, J., Nimchuk, Z., Mudgett, M. B., Elowe, S., Gee, C. L., Liu, L., Daly, R. J., Manning, G., Babon, J. J. and Lucet, I. S. (2014). A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem J 457(2): 323-334.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lucet, I. S., Hildebrand, J. M., Czabotar, P. E., Zhang, J., Nicola, N. A., Silke, J., Babon, J. J. and Murphy, J. M. (2014). Determination of Pseudokinase-ligand Interaction by a Fluorescence-based Thermal Shift Assay. Bio-protocol 4(11): e1135. DOI: 10.21769/BioProtoc.1135.
Category
Biochemistry > Protein > Fluorescence
Biochemistry > Protein > Interaction > Protein-ligand interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









