- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Arabidopsis thaliana Embryo Sac Mitochondrial Membrane Potential Stain
Published: Vol 4, Iss 10, May 20, 2014 DOI: 10.21769/BioProtoc.1128 Views: 11456
Reviewed by: Beery YaakovSamik BhattacharyaRu Zhang

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2322 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1690 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 756 Views
Abstract
The aim of this experiment is to study mitochondrial functional status in Arabidopsis embryo sacs using the membrane potential indicator JC-1. Changes in the membrane potential are presumed to be due to the opening of the mitochondrial permeability transition pore (MPTP), allowing passage of ions and small molecules. The resulting equilibrium of ions leads in turn to the decoupling of the respiratory chain and the release of cytochrome c into the cytosol, a distinctive feature of the early stages of programmed cell death.
JC-1 is a lipophilic dye that can selectively enter into mitochondria and reversibly change color from green to red as the membrane potential increases. In healthy cells with high mitochondrial potential, JC-1 spontaneously forms complexes with intense red fluorescence. On the other hand, in mitochondria with low mitochondrial potential, JC-1 remains in the monomeric form, which exhibits only green fluorescence (Martin et al., 2013; Hauser et al., 2006).
This protocol could be used in isolated mitochondria, and in a variety of cell types and different tissues of plants and other organism.
Materials and Reagents
- Flowers at different developmental stages from an Arabidopsis inflorescence
- JC-1 dye (Life Technologies, Molecular Probes®, catalog number: T3168 )
- DMSO 99.9% (Sigma-Aldrich, catalog number: D8418 )
- Stock solution (10 mg/ml of JC-1 in DMSO)
- Working solution (10 µg/ml of JC-1 in buffer A)
- Buffer A (20 mM HEPES buffer, pH 7.2) (Sigma-Aldrich, catalog number: H3375 ) (see Recipes)
Equipment
- Confocal microscope (Nikon Eclipse C1 Plus Confocal microscope, using EZ-C1 3.80 imaging software and Ti-Control)
- Dissecting microscope (Nikon Corporation, model: SMZ800 )
- Coverslip (18 x 18 mm)
- Microscopic slide (26 x 76 mm)
- 1 ml insulin syringe with the 0.3 x 13 mm needle (BD)
- Needle point tweezers
Software
- NIH ImageJ software 1.47 for Windows (http://rsb.info.nih.gov/ij/)
Procedure
- Pistils isolation (Figure 1, A-E).
Using a pair of tweezers, take flowers, at different developmental stages from an Arabidopsis inflorescence (Alvarez-Buylla et al., 2010). On a microscopic slide and under a dissecting microscope, remove sepals, petals, anthers and stigma using the needles of two 1 ml insulin syringes (0.3 x 13 mm) and make longitudinal superficial cuts on pistils at each side of the septum (Figure 1).
- Pistils stain (Figure 1, F-G).
- Submerge the pistils in 100 µl of a solution containing 10 µg/ml of JC-1 in buffer A.
- Incubate for 30 min at room temperature without shaking. Protect from light, as the dye is photosensitive.
- Gently wash the pistils three times with buffer A.
- Submerge the pistils in 100 µl of a solution containing 10 µg/ml of JC-1 in buffer A.
- Sample preparation for microscopy (Figure 1, H-L).
- On a microscopic slide under a dissecting microscope, use the needles of two 1 ml insulin syringes (0.3 x 13 mm) to dissect the pistils exposing the ovules
- Add a drop of buffer A and cover the sample with a coverslip.
- Immediately observe under a confocal microscope. The intensities of green (excitation/emission wavelength = 485/538 nm) and red (excitation/emission wavelength = 485/590 nm) are analyzed.
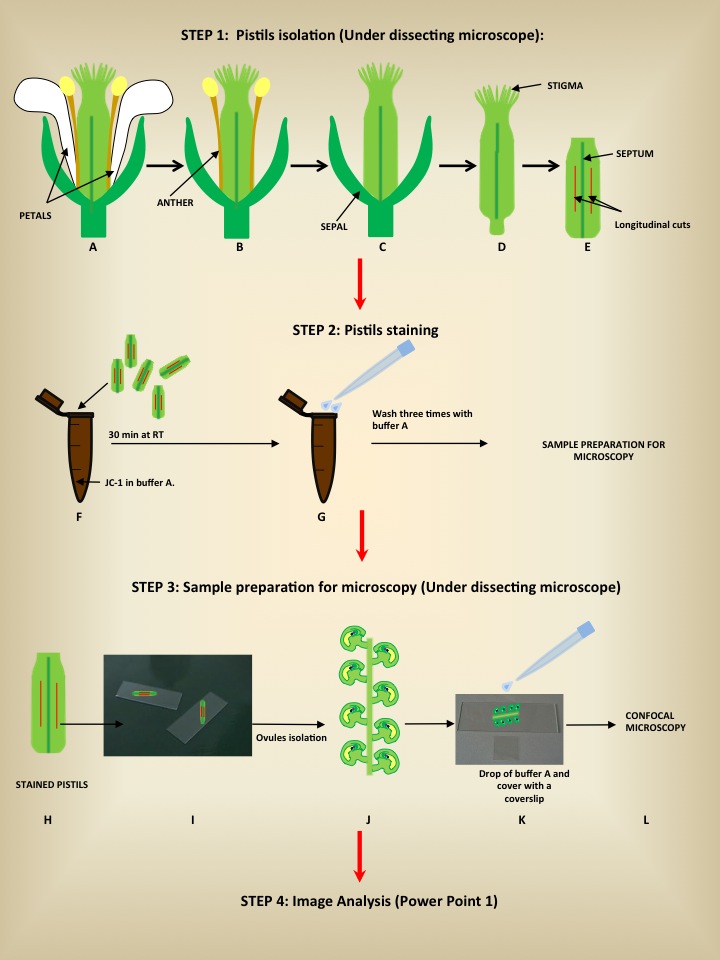
Figure 1. Schematic illustration showing the steps required for embryo sac mitochondrial staining
- On a microscopic slide under a dissecting microscope, use the needles of two 1 ml insulin syringes (0.3 x 13 mm) to dissect the pistils exposing the ovules
- Image analysis (Power Point 1). The ratio of red to green fluorescence of JC-1 images is calculated using NIH ImageJ software.
- The outline of each embryo sac is delimited using the freehand tool to create a region of interest (ROI) and saved using the ROI manager tool (Analyze>tools>ROI manager>Add).
- In the “Analyze” menu, select “set measurements” and click on area and “mean gray value”.
- Using the image Menu, select “color” and then “split channels”.
- Close the image in the blue channel. On the green channel, go to the analyze>tools>ROI manager and select the ROI added before. The ROI will appear on the image. Still in the ROI manager go to more>multi Measure. The result will appear in a new window.
- Repeat this step with the image in the red channel.
- Copy the results and paste them in an excel worksheet.
- Calculate the red to green fluorescence ratio for each ROI.
- The outline of each embryo sac is delimited using the freehand tool to create a region of interest (ROI) and saved using the ROI manager tool (Analyze>tools>ROI manager>Add).
Recipes
- Buffer A
20 mM HEPES buffer (pH 7.2)
For 1 L of 1 M HEPES buffer:
Dissolve 238.3 g HEPES (free acid) in 500 ml of ddH2O
Stir while adjusting the pH 7.2 with 0.5 N NaOH
Bring up the volume to 1 L with ddH2O to prepare 1 L of 20 mM HEPES buffer (Buffer A)
Add 20 ml of 1 M HEPES buffer in 980 ml of ddH2O
Acknowledgments
This protocol was adapted from Hauser et al. (2006). This work was supported by The Howard Hughes Medical Institute (HHMI), National Scientific and Technical Research Council (CONICET), National Agency for Promotion of Science and Technology (AGENCIA) and National University of Mar del Plata (UNMdP). We are grateful to the Editorial Committee of Bio-protocol for kindly inviting us to write this protocol.
References
- Alvarez-Buylla, E. R., Benitez, M., Corvera-Poire, A., Chaos Cador, A., de Folter, S., Gamboa de Buen, A., Garay-Arroyo, A., Garcia-Ponce, B., Jaimes-Miranda, F., Perez-Ruiz, R. V., Pineyro-Nelson, A. and Sanchez-Corrales, Y. E. (2010). Flower development. Arabidopsis Book 8: e0127.
- Hauser, B. A., Sun, K., Oppenheimer, D. G. and Sage, T. L. (2006). Changes in mitochondrial membrane potential and accumulation of reactive oxygen species precede ultrastructural changes during ovule abortion. Planta 223(3): 492-499.
- Martin, M. V., Fiol, D. F., Sundaresan, V., Zabaleta, E. J. and Pagnussat, G. C. (2013). oiwa, a female gametophytic mutant impaired in a mitochondrial manganese-superoxide dismutase, reveals crucial roles for reactive oxygen species during embryo sac development and fertilization in Arabidopsis. Plant Cell 25(5): 1573-1591.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Martin, M. V., Fiol, D. F., Zabaleta, E. J. and Pagnussat, G. C. (2014). Arabidopsis thaliana Embryo Sac Mitochondrial Membrane Potential Stain. Bio-protocol 4(10): e1128. DOI: 10.21769/BioProtoc.1128.
Category
Plant Science > Plant cell biology > Cell imaging
Plant Science > Plant developmental biology > General
Cell Biology > Cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link