- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Enterovirus 71 Virus Propagation and Purification
Published: Vol 4, Iss 9, May 5, 2014 DOI: 10.21769/BioProtoc.1117 Views: 14210
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

General Maintenance and Reactivation of iSLK Cell Lines
Ariana C. Calderón-Zavala [...] Ekaterina E. Heldwein
Jun 5, 2025 1896 Views

Inducible HIV-1 Reservoir Reduction Assay (HIVRRA), a Fast and Sensitive Assay to Test Cytotoxicity and Potency of Cure Strategies to Reduce the Replication-Competent HIV-1 Reservoir in Ex Vivo PBMCs
Jade Jansen [...] Neeltje A. Kootstra
Jul 20, 2025 2460 Views

Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 3038 Views
Abstract
Since its discovery in 1969, enterovirus 71 (EV71) has emerged as a serious worldwide health threat. This member of the picornavirus family causes hand, foot, and mouth disease, and also has the capacity to invade the central nervous system to cause severe disease and death. This is the propagation and purification procedure to produce infectious virion.
Materials and Reagents
- Hela cells
- EV71 inoculum (ATCC, www.atcc.org)
- DMEM (Sigma-Aldrich, catalog number: SH3002201 )
- Fetal Bovine Serum (Thermo Fisher Scientific, catalog number: SH3039603 )
- Polyethylene glycol (PEG) 8000 (Thermo Fisher Scientific, catalog number: P156-3 )
- Tris-HCl
- Magnesium chloride (MgCl2)
- 0.5 M NaCl
- 0.05 mg/ml DNase (Thermo Fisher Scientific, catalog number: NC9709009 )
- 0.1 M EDTA (pH=8.0)
- Ammonium hydroxide (Sigma-Aldrich, catalog number: A5132-5 Kg )
- Potassium tartrate (Sigma-Aldrich, catalog number: 25516-500 g )
- 100 kD cutoff spin column with 4 ml capacity (Millipore, catalog number: UFC810096 )
- 30% sucrose-cushion in purification buffer
- Purification buffer (see Recipes)
Equipment
- Cell Stacks (1 10-stack and 1 2-stack) (Thermo Fisher Scientific, catalog numbers: 12-567-303 and 12-567-301 )
- Centrifuge
- SLA1500 rotor
- 50.2ti rotor
- 26.3 ml red-capped Beckman tubes (Beckman Coulter, catalog number: 355618 )
- SW41 rotor
- Beckman Ultra-Clear centrifuge tubes (14 x 89 mm) (Beckman Coulter, catalog number: 344059 )
- Chemistry ring stand with clamp
- Small light source
- Microcentrifuge
Procedure
- 90-95% confluent Hela cell monolayers (passage number not exceeding 30) grown in cell stacks (1 10-stack and 1 2-stack) were infected with EV71 strain 1095/Shiga at an MOI of 0.1 in culture with 600 ml of DMEM (500 ml in 10-stack and 100 ml in 2-stack). Virus was allowed to attach and infect for one hour at 37 °C. 600 ml of DMEM supplemented with 5% fetal bovine serum (500 ml in 10-stack and 100 ml in 2-stack) was added to achieve a final volume of 1,200 ml total (1,000 ml in 10-stack and 200 ml in 2-stack). Infections were allowed to continue at 37 °C.
- Cells and media were harvested 24 h post infection, (or as soon as cytopathic effects were obvious) and subjected to three freeze-thaw cycles. To remove cell debris, the lysate was centrifuged at 13,000 rpm in a SLA1500 rotor at 4 °C for 15 min. To the lysate, PEG 8000 was added to provide a final concentration of 8% and NaCl, was added to provide a final concentration of 0.5 M. Virus was precipitated overnight at 4 °C with stirring and then centrifuged in a SLA1500 rotor (4 °C, 13,000 rpm, 45 min).
- Pellets were resuspended in 10 ml purification buffer, bottles were rinsed with 5 ml purification buffer, and 0.05 mg/ml DNase was added (do not exceed total buffer volume of 20 ml). The suspension was incubated at room temperature for 10 min with gentle rocking.
- After incubation, 0.1 M EDTA (pH=8.0) was added (10% total volume), the pH was adjusted using ammonium hydroxide, and supernatant was cleared by low speed centrifugation (4,000 rpm, 5 min, 4 °C).
- The supernatant was transferred to a red-capped Beckman tube. 2 ml of 30% sucrose in purification buffer was pipetted directly underneath the virus-containing supernatant. The virus was then pelleted through the sucrose cushion (50.2ti rotor, 48,000 rpm, 4 °C, 2 h).
- The pellet was resuspended in 2 ml of purification buffer, tube rinsed with an additional 1 ml of purification buffer, centrifuged at 4,000 rpm for 5 min to remove any remaining cellular debris, and supernatant applied to a 10-35% potassium tartrate (K-Tartrate) step gradient in the Ultra-clear centrifuge tubes for final purification by ultracentrifugation (36,000 rpm, 4 °C, 2 h, SW41 rotor).
- The centrifuge tube was immobilized in a clamp on a ring stand above a small light source (Figure 1). Other light sources were extinguished to visualize the two distinct bands of virus, which were collected by side puncture and diluted in purification buffer to reach a final volume of 4 ml. The 4 ml sample was placed into a 100 kD cutoff spin column and the volume was reduced to about 50-100 microliters by centrifugation at 4,000 rpm. The sample was diluted again with purification buffer to return the volume to 4 ml and then centrifuged to reduce volume to 50-100 microliters. This process of washing the sample to remove tartrate and replace it with purification buffer was repeated in three more wash steps (four in total).
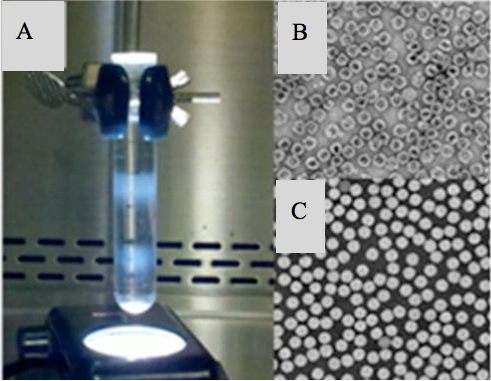
Figure 1. A) Two bands of virus in the tartrate gradient are visible when a small light source is placed beneath the centrifuge tube. B) The upper band contains empty capsids, which have intact VP0 whereas the lower band C) contains native infectious virus. As assayed by negative stain TEM (shown) and PAGE gel (data not shown). - The concentration of the final 100 microliters of virus was determined by spectrometry reading (procapsid read at wavelength of 280 nm and native virus read at a wavelength of 260 nm).
- As previously reported the upper band consisted of procapsid, characterized by the presence of uncleaved VP0 and lack of genomic material. The lower band consisted of native virus comprised of VP2 and VP4, with packaged genome. Each species was diluted to a concentration of 0.1 mg/ml for storage. Procapsid is stored at 4 °C and native virus is stored at -80 °C.
Recipes
- Purification buffer
10 mM Tris-HCl
200 mM NaCl
50 mM MgCl2
pH=7.5 (if initial pH is < 7.5, adjust with 1 M NaOH, if initial pH is > 7.5, adjust with 1 M HCl)
Acknowledgments
This work was funded in part by the Pennsylvania Department of Health Tobacco CURE Funds and the Junior Faculty Research Scholar Max Lang Award from the Pennsylvania State University College of Medicine. This protocol was previously described in Shingler et al. (2013).
Competing interests
The authors declare no conflict of interest or competing interest.
References
- Cifuente, J. O., Lee, H., Yoder, J. D., Shingler, K. L., Carnegie, M. S., Yoder, J. L., Ashley, R. E., Makhov, A. M., Conway, J. F. and Hafenstein, S. (2013). Structures of the procapsid and mature virion of enterovirus 71 strain 1095. J Virol 87(13): 7637-7645.
- Shingler, K. L., Yoder, J. L., Carnegie, M. S., Ashley, R. E., Makhov, A. M., Conway, J. F. and Hafenstein, S. (2013). The enterovirus 71 A-particle forms a gateway to allow genome release: a cryoEM study of picornavirus uncoating. PLoS Pathog 9(3): e1003240.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Shingler, K. L., Organtini, L. J. and Hafenstein, S. (2014). Enterovirus 71 Virus Propagation and Purification. Bio-protocol 4(9): e1117. DOI: 10.21769/BioProtoc.1117.
Category
Microbiology > Microbial cell biology > Cell isolation and culture
Microbiology > Microbe-host interactions > Virus
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












