- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation and Immortalization of Fibroblasts from Different Tumoral Stages
Published: Vol 4, Iss 7, Apr 5, 2014 DOI: 10.21769/BioProtoc.1097 Views: 20324
Reviewed by: Lin FangFanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Mammary Epithelial Cells and Fibroblasts From Mouse Tumor
Shiva Kazerounian
Feb 20, 2014 20828 Views

In vivo Tumor Growth and Spontaneous Metastasis Assays Using A549 Lung Cancer Cells
Lei Qi [...] Min Chen
Apr 5, 2020 8839 Views

Generation and Maintenance of Patient-Derived Endometrial Cancer Organoids
Mali Barbi [...] Semir Beyaz
Oct 20, 2024 2061 Views
Abstract
Tumour microenvironment and cancer-associated fibroblasts in particular exhibit tumour promoting abilities that are not present in their normal counterparts (Calvo et al., 2013; Hanahan and Coussens, 2012). Therefore, functional and molecular characterization of the modifications occurring in fibroblasts during tumour progression is essential to fully understand their role in tumour progression. Previous studies have addressed this issue using human fibroblasts and comparing normal and adjacent fibroblasts to tumour-associated fibroblasts (Kalluri and Zeisberg, 2006). However, these studies are hampered by the intrinsic variability of human samples (e.g. pairing, age, genomic landscape, etc). In order to overcome these issues, we used a fully characterised mouse breast cancer model, MMTV-PyMT (Guy et al., 1992; Lin et al., 2003). MMTV-PyMT transgenic mice express the Polyoma Virus middle T antigen under the direction of the mouse mammary tumor virus promoter/enhancer. This is a multifocal luminal breast cancer model that goes through well defined and characterised stages (namely, hyperplasia, adenoma, carcinoma and invasive carcinoma). Interestingly, this model has a 100% incidence, is very desmoplastic (presenting high concentration of fibroblasts) and gives raise to spontaneous metastasis in the lung with 80-94% incidence. Importantly, at least for the inguinal mammary glands (glands 4 and 9), the different tumoral stages are well correlated to the age of the mouse: hyperplasia arising at 6 weeks of age, adenoma between 6-8 weeks of age, carcinoma and invasive carcinoma from 8 weeks onwards. This model allowed us to confidently isolate fibroblasts from different tumoral stages and carefully characterise their functional and molecular properties (Calvo et al., 2013).
Materials and Reagents
- FVB/n MMTV-PyMT females between 4 and 14 weeks of age and MMTV-PyMT negative siblings for isolation of normal mammary gland fibroblasts (NFs) (The Jackson Laboratory, catalog number: 00 2374 )
- Phoenix-Eco packing cells (ATCC, catalog number: CRL-3214 ) (read Note 6 for more information)
- pBabe-HPV-E6-puromycin plasmid (or immortalization plasmid of choice, see Note 6)
Note: This was generated in our laboratory but there are alternatives available (pLenti-puro-HPV-16 E6/E7; Applied Biological Materials, catalog number: G268 ). - Dulbecco’s Modified Eagle’s Medium (DMEM) (high glucose with stable L-glutamine) (Life Technologies, Gibco®, catalog number: 41966-029 )
- 50x sodium butyrate (500 mM)
- Polybrene® (1,5-dimethyl-1,5-diazaundecamethylene polymethobromide, hexadimethrine bromide) (Sigma-Aldrich, catalog number: AL-118 ) (1000x solution at 2 mg/ml)
- Insulin-Transferrin-Selenium (ITS) solution (Life Technologies, catalog number: 51300-044 ).
- Calcium phosphate transfection kit (ProFection® Mammalian Transfection System) (Promega Corporation, catalog number: E1200 )
- Fetal Bovine Serum (FBS)
- Phosphate-buffered saline (PBS)
- PBS without Ca2+ and Mg2+
- Penicillin/Streptomycin
- EDTA (1 mM in PBS without Ca2+ and Mg2+)
- 10% formalin solution, neutral-buffered (10% NBF)
- 100x Collagenase/dispase (100 mg/ml) (Roche Diagnostics, catalog number: 11097113001 )
- 1000x puromycin (2 mg/ml) (Sigma-Aldrich, catalog number: P9620 )
- 1000x DNase I (10 mg/ml) (Sigma-Aldrich, catalog number: D4513 )
- Trypsin/EDTA
- Virkon (DuPont Rely+On Virkon, catalog number: 1235-8667 )
- Complete media (see Recipes)
Equipment
- 0.22 µm, 0.45 µm, 3 mm filters
- 20 mm coverslip
- Tissue culture plastic-ware
- 37 °C shaker
- Tissue culture laminar-flow hood
- 10 cm tissue culture dish
- Centrifuge
- 37 °C, 5% CO2, cell culture incubator
- Scalpels
- Surgical scissors
- Dissecting instruments
Procedure
- Before starting
- Please read Notes section for guidance.
- Obtain MMTV-PyMT positive females between 5 and 14 week of age.
Note: Correlation between age and tumoral stage may vary in other tumor models. Read Notes section for more information. - Make sure you have all the materials, reagents and equipment.
- Please read Notes section for guidance.
- Dissecting samples – tumor material
- Mammary tissue from the 4th and 9th inguinal glands is dissected using surgical instruments. Tissue is kept in ice-cold PBS until further use.
Note: For normal mammary gland isolation, mammary tissue of MMTV-PyMT negative females of 10 weeks of age is dissected. For isolation of hyperplastic tumors, mammary tissue of MMTV-PyMT positive females of 6 weeks of age is dissected. For isolation of adenoma, carcinoma, and invasive carcinoma, mammary tissue of MMTV-PyMT positive females of 7, 8 and 10 weeks of age, respectively, is dissected (Figure 1). These time points may vary. For more information about the determination of the correct age and tumoural stage, please read Note 2 at the end of the protocol. - Samples are moved into a laminar flow tissue culture hood under sterile conditions.
- Tissue samples are carefully analysed checking for signs indicative of their tumoral stage. As a rough guide, normal tissue appears fatty and soft. Hyperplastic tissue appears considerably enlarged but still fatty and soft. Adenoma, carcinoma, and invasive carcinoma appear enlarged, stiff and multimodular to variable degrees (Figure 1).
- Isolate single modules on each tumor (2-3 per tumour is recommended.) before proceeding to the next stage.
Note: As a result of the multimodular aspect of this type of tumour, special care must be taken in order to use and analyse only single modules. This is highly important if you want to make sure that you are isolating cells from a specific stage as individual modules may have progressed to different tumoral stages. The size of the modules varies with tumour size, age, localization and between individual mice. Expert pathological advice is recommended (see Notes 2-3). - For each single module, take one part for histopathology analysis and one part for isolation of fibroblasts. For histopathological analysis, the sample is fixed in 10% NBF overnight at 4 °C. See Figure 1 for representative hematoxylin and eosin staining of tissues.
Note: Histopathological analysis procedures will not be discussed here. Please refer to other protocols. - For isolation of fibroblasts, the rest of the sample is transferred to a 10 cm tissue culture dish and processed into smaller pieces using a scalpel and/or surgical scissors.
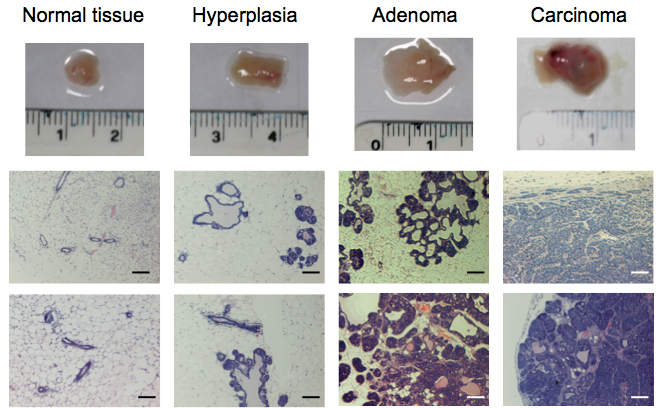
Figure 1. Representative tissue samples and hematoxylin and eosin staining of tissues from normal FVB/n mammary glands and progressive breast tumoural stages (hyperplasia, adenoma and carcinoma) from the MMTV-PyMT model. Scale bars represent 100 µm.
- Mammary tissue from the 4th and 9th inguinal glands is dissected using surgical instruments. Tissue is kept in ice-cold PBS until further use.
- Processing normal mammary tissue samples
- For normal mammary tissue, the smaller tissue pieces are placed into dishes where they are compressed under a 20 mm coverslip to prevent the fatty tissue floating.
Note: If normal mammary tissue is processed using the collagenase/disparse approach (see below), very low numbers of fibroblasts are isolated. - Fresh complete media DMEM is then added slowly to prevent samples from floating.
Note: Samples need to be in contact with the plastic and the coverslip in order for the fibroblasts to come out of the sample. - Media is replaced daily with fresh complete media for 7 days.
Note: After 7 days, fibroblasts have come out of the tissue into the coverslip and the dish and can be transferred into a new dish. - Original tissue is removed. Coverslips are transferred to a new dish (side with cells facing up).
- The original dish and the dish with coverslips are washed once with PBS and fibroblasts trypsinized using Trypsin/EDTA for 10 min at 37 °C.
- Cells are resuspended and seeded into a new dish.
Note: Cell numbers can be expanded minimally, always taking into account that primary cells enter senescence after prolonged culture in vitro. - Cells are now ready for immortalization (starting at step E30).
- For normal mammary tissue, the smaller tissue pieces are placed into dishes where they are compressed under a 20 mm coverslip to prevent the fatty tissue floating.
- Processing mammary hyperplasia, adenoma and carcinoma tissue samples
- For hyperplasia, adenoma and carcinoma samples smaller tissue pieces are placed into dishes where they are further disgregatted using scalpels.
Note: Alternatively, for extremely fatty hyperplasia samples normal tissue isolation approach (steps C10-16) can be used. - Collagenase/dispase solution (100 mg/ml) is diluted 1:100 in sterile PBS Mg2+ Ca2+ free and filtered before use with 22 µm filter (final concentration 1 mg/ml).
Note: Collagenase/dispase solution is a tissue dissociation solution that degredes collagen and reticular fibers without affecting the integrity of the plasma membrane. - Disgregatted tissues are placed in a sterile tube with the appropriate amount of collagenase/dispase. Pipette up and down several times to help disaggregate. Samples are then incubated for 60 min at 37 °C and 180 rpm in an orbital shaker.
Note: Generally use 2-3 ml per sample (250-500 mg of tissue). Adjust to sample size. - One volume of PBS 1 mM EDTA is added to each sample. Pass it through the pipette several times.
Note: EDTA stops the protease reaction as it is a collagenase inhibitor. - Samples are filtered using the 3 mm filters.
Note: This step removes undigested pieces of tissue. Alternative methods are also available (rubber syringe plunger, etc). - Samples are centrifuged at 100 x g for 5 min at room temperature (RT).
- Supernatant is discarded. Pellet containing cells is washed with complete media and centrifuged at 100 x g for 5 min at RT again.
- Supernatant is discarded. Pellet containing cells is incubated for 10 min at RT with complete media plus DNase 1 (final concentration 10 µg/ml).
Note: Often as a result of cell damage, DNA leaks into the dissociation medium increasing viscosity and causing handling problems. Purified DNase is included in cell isolation procedures to digest the nucleic acids without damaging the intact cells. - Samples are centrifuged at 100 x g for 5 min at room temperature (RT).
- Repeat step D23 twice.
- Media is removed and cells are resuspended in complete media and seeded in a dish. Place samples in the incubator.
- After 30 min, media in all samples is replaced with fresh complete media and place back in the incubator.
Note: After 30 min, most of the fibroblasts would have adhered to the dish, whilst other cell types will remain in suspension. Using this tip will help you enrich for the fibroblastic population. - After 2-3 days, isolated cells can be enriched for the fibroblastic population using the enhanced adherence to plastic of fibroblasts.
- Media is removed.
- Cells are washed with PBS once.
- Cells are covered with Trypsin/EDTA and placed in the incubator for 10 min.
- Cells are resuspended in complete media, seeded into a new dish and placed back in the incubator for 30 min.
- Media is replaced with fresh new complete media.
- Media is removed.
- For hyperplasia, adenoma and carcinoma samples smaller tissue pieces are placed into dishes where they are further disgregatted using scalpels.
- Immortalizing primary fibroblasts from different tumoral stages
- Read Notes 4-5 before start.
- Growing Phoenix-ECO cells are seeded to reach 80% confluency by the time of transfection (see Note 6).
Note: Phoenix-Eco cells are used to produce ecotropic viruses to infect mouse fibroblasts. To immortalize human fibroblasts, please use alternative approaches (see Notes 4 and 6). Optimization of volumes and titration of retroviruses is highly recommended but will not be discussed here. Using easy-to-spot positive controls (e.g. transfection of retroviral plasmids encoding fluorescent proteins) is also recommended. This allows for rapid evaluation of your infection efficiency (e.g. scoring for fluorescently positive cells). - Once they reach 80% confluency, Phoenix-Eco cells are transfected using the calcium phosphate protocol. Usually for 10 cm plate.
- Replace media with complete media 2 h before transfection.
- In Tube A, mix pBabe-HPV-E6-puromycin (10 µg), 2 M CaCl2 (67 µl) and Nuclease-free water (up to 500 µl).
- In Tube B, add 500 µl of 2x Hepes.
- While bubbling the content in Tube A, slowly add content of Tube B into it.
- Leave 30 min at RT.
- Add transfection mix dropwise into Phoenix-Eco cells.
- Replace media with complete media 2 h before transfection.
- After 18 h, in order to burst transcription, Sodium Butyrate is added to the cells (final concentration 10 mM).
- After 4 h, media is replaced with fresh complete media.
- After 48-72 h, media containing viruses is collected and filtered through a 0.45 µm filter. Virus can be aliquoted and stored at -80 °C until used.
Note: All used material must be left o/n in virkon before disposal (see Note 5). - Primary fibroblasts are infected with HPV-E6-puromycin retroviruses.
- Media on fibroblast cultures is removed.
- The virus-containing media is mixed 1:1 with complete fresh media and the resulting solution added to the fibroblasts.
- Polybrene (final concentration 2 µg/ml) is also added to your cells.
Note: Infection efficiency may increase if receptor cells are in suspension (e.g. after trypsinization). Polybrene (hexadimethrine bromide) is a cationic polymer used to increase the efficiency of infection of certain cells with a retrovirus in cell culture.
- Media on fibroblast cultures is removed.
- After 5 h, media is replaced with fresh media.
Note: Alternatively, media can be replaced after 18 h. - After 24-48 h, infected cells are selected by addition of puromycin (2 µg/ml final concentration) in the complete culture media.
Note: Puromycin subsceptibility varies depending on the cell type. Final concentration needs to be optimized prior to final use. - After 7-10 days, resistant clones expressing HPV-E6 (i.e. immortalized cell lines) are visible. Clones or pools can be expanded in culture of frozen down for subsequent use.
Note: Eventually, characterization of the nature of the isolated and immortalized populations must be performed. This can be performed by analysis of expression of certain markers by immunofluorescence, FACS, etc. Recommended fibroblast markers are CD140A/PDGFRA, CD90/Thy-1, Vimentin, and Fibronectin. Fibroblasts should also be negative for immune cell markers (CD45 negative), endothelial markers (CD31 negative) and epithelial markers (E-Cadherin, EpCAM negative). For cancer-associated fibroblast markers we recommend αSMA, FAP and FSP1/S100A4.
- Read Notes 4-5 before start.
Notes
- This procedure works exceptionally well because it is optimized for this tumour model. Importantly, MMTV-PyMT breast cancer cells do not grow on tissue culture plastic dishes. On the other hand, fibroblasts grow very well. Therefore, just by culturing your extracts in plastic, you are enriching for the fibroblast population and any contamination derived from cancer cells is avoided. However, this procedure does not work if established cancer cell lines are used, as those cells are adapted to grow in plastic and will eventually overtake the fibroblast population. In order to prevent this, other approaches need to be designed (FACS sorting, use of suicide genes in cancer cells, etc).
- This procedure needs a well-characterized model and much familiarization with the different tumoral stages of its development. Therefore, it is essential that, prior to its application, the researcher gets familiar with it. Expert pathological evaluation is essential. We recommend thorough pathological analysis of multiple tumours of different ages, sizes, aspects and sites. This will eventually allow the researcher to confidently predict the stage of each tumour. Even then, retrospective pathological analysis of the tumoral stage of the tumours used for fibroblast isolation must always be performed.
- Histopathology and immunocytochemistry approaches will not be discussed in this protocol. Tumour progression is highly variable depending on type, tissue, mouse strain, etc. In order to characterise each tumoral stage, consult a pathologist to get advice about the best approaches, techniques and markers to clearly define each stage.
- Primary cells and specially fibroblasts can be maintained in culture for few passages before they become senescent. Senescence induces a pletora of transcriptional and functional changes that may affect profoundly the phenotypes under study. Therefore, if long term culture is desired, immortalization is necessary. Ideally, primary and immortalized cultures should be compared in order to check that the immortalization procedure has not induced any change in the phenotypes under study. For immortalizing mouse fibroblasts, we express the human papillomavirus “early gene” E6 that prevents replicative senescence by suppressing p53 activity. As a result, other “off-target” consequences in specific functions and signalling pathways are to be expected. For cells that are most affected by telomere length (e.g. human), an alternative approach is recommended. In those cases, cell immortalization can be achieved by expression of Telomerase Reverse Transcriptase protein (hTERT). This protein is inactive in most somatic cells, but when hTERT is exogenously expressed, the cells are able to maintain sufficient telomere lengths to avoid replicative senescence. Analysis of several telomerase-immortalized cell lines has verified that the cells maintain a stable genotype and retain critical phenotypic markers.
- Immortalization is achieved by retroviral infection of target fibroblasts. Thus, handling and disposal of viruses and contaminated material must be always performed in accordance with you institution regulations.
- Phoenix-ECO were originally established in Garry P. Nolan lab (Stanford University, USA) and are available at ATCC (Pear et al., 1993). They are a retrovirus producer line for the generation of helper free ecotropic retroviruses. The line is based on the 293T cell line (a human embryonic kidney line transformed with adenovirus E1a and carrying a temperature sensitive T antigen co-selected with neomycin). This cell line is highly transfectable with either calcium phosphate mediated transfection or lipid-based transfection protocols. The line was created by placing into 293T cells constructs capable of producing gag-pol, and envelope protein for ecotropic viruses. Phoenix-Ampho is a similar cell line that instead produces amphotropic retroviruses.
Recipes
- Complete media (always to be used warm 37 °C)
Dulbecco’s Modified Eagle’s Medium (DMEM) high glucose with stable L-glutamine
1% Insulin-Transferrin-Selenium solution (ITS)
10% Fetal Bovine Serum (FBS)
Penicillin/Streptomycin
Acknowledgments
This protocol is an extended version of the one described in Calvo et al. (2013). FC, SH and ES were supported by a Cancer Research UK grant CRUK_A5317. We thank lab members for help and advice throughout this work.
References
- Calvo, F., Ege, N., Grande-Garcia, A., Hooper, S., Jenkins, R. P., Chaudhry, S. I., Harrington, K., Williamson, P., Moeendarbary, E., Charras, G. and Sahai, E. (2013). Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol 15(6): 637-646.
- Guy, C., Cardiff, R. and Muller, W. (1992). Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cellr Biol 12(3): 954-961.
- Hanahan, D. and Coussens, L. M. (2012). Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21(3): 309-322.
- Kalluri, R. and Zeisberg, M. (2006). Fibroblasts in cancer. Nat Rev Cancer 6(5): 392-401.
- Lin, E. Y., Jones, J. G., Li, P., Zhu, L., Whitney, K. D., Muller, W. J. and Pollard, J. W. (2003). Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol 163(5): 2113-2126.
- Pear, W. S., Nolan, G. P., Scott, M. L. and Baltimore, D. (1993). Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A 90(18): 8392-8396.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Calvo, F., Hooper, S. and Sahai, E. (2014). Isolation and Immortalization of Fibroblasts from Different Tumoral Stages. Bio-protocol 4(7): e1097. DOI: 10.21769/BioProtoc.1097.
Category
Cancer Biology > General technique > Animal models > Cell isolation and culture
Cancer Biology > Invasion & metastasis > Tumor microenvironment > Tissue isolation
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










