- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
UPF1 RNA Immunoprecipitation from Mini-μ Construct–expressing Cells
Published: Vol 4, Iss 7, Apr 5, 2014 DOI: 10.21769/BioProtoc.1086 Views: 11467
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Individual-nucleotide-resolution UV Cross-linking and Immunoprecipitation (iCLIP) of UPF1
David Zünd and Oliver Mühlemann
Apr 5, 2014 15084 Views

RNA-binding Protein Immunoprecipitation (RIP) to Examine AUF1 Binding to Senescence-Associated Secretory Phenotype (SASP) Factor mRNA
Elise Alspach and Sheila A. Stewart
May 20, 2015 15258 Views

Assessing Self-interaction of Mammalian Nuclear Proteins by Co-immunoprecipitation
Claudia Cattoglio [...] Anders S. Hansen
Feb 20, 2020 9471 Views
Abstract
UPF1, an RNA helicase and a core factor of nonsense-mediated mRNA decay (NMD), interacts with RNA independently of the sequence context. To investigate the influence of translation on the association of UPF1 with specific reporter transcripts, UPF1 RNA immunoprecipitations (RIPs) are performed from Hela cells that either express a normally translated immunoglobulin-µ (Ig-µ) reporter (mini µ) or a version with a stable stem loop in the 5' UTR (SL mini µ) that efficiently inhibit translation initiation (Zund et al., 2013). Both the cloning of the SL mini µ reporter construct and the UPF1 RIP experiment are described in detail.
Materials and Reagents
- Hela cells
- XL-10 Gold Ultracompetent Cells (Agilent Technologies, Stratagene, catalog number: 200315 )
- Dulbecco’s Modified Eagle Media (DMEM) (powder, high glucose) (Life Technologies, catalog number: 52100-039 )
- MQ-water (pure water from the ELGA system)
- Fetal Calf Serum (FCS) (BioConcept, Amimed, catalog number: 2-01F30-I )
- Penicillin-Streptomycin solution (P/S) (1 unit/ml) (BioConcept, Amimed, catalog number: 4-01F00-H )
- Trypsin-EDTA (T/E) (BioConcept, Amimed, catalog number: 5-51F00-H )
- DreamFectTM (OZ Biosciences, catalog number: DF45000 )
- KpnI (New England Biolabs, catalog number: R0142S )
- SL Oligo 1 (Microsynth AG)
Oligo sequence: 5'-CGGGTTCCGTCCAAGCACTGTTGAAGCAGGAAACCCGGGTTGCTAGTCGATCGACTAG-CAACCCGGGTTTCCTGCTTCAACAGTGCTTGGACGGAACCCCGATCGTAC-3' - SL Oligo 2 (Microsynth AG)
Oligo sequence: 5'-GATCGGGGTTCCGTCCAAGCACTGTTGAAGCAGGAAACCCGGGTTGCTAGTCGATCG-ACTAGCAACCCGGGTTTCCTGCTTCAACAGTGCTTGGACGGAACCCGGTAC-3' - T4 DNA Ligase (New England Biolabs, catalog number: M0202S )
- T4 Polynucleotide Kinase (PNK) (New England Biolabs, catalog number: M0201L )
- Alkaline phosphatase calf intestinal (CIAP) (Promega Corporation, catalog number: M182A )
- 100x HaltTM Protease Inhibitor Cocktail (Thermo Fisher Scientific, catalog number: 1861279 )
- RiboLock RI RNase Inhibitor (40 U/µl) (Thermo Fisher Scientific, catalog number: EO0381 )
- RNase I (cloned, 100 U/µl) (Life Technologies, Ambion®, catalog number: AM2294 )
- Turbo DNase (Life Technologies, Ambion®, catalog number: AM2238 )
- Glycogen for molecular biology (Roche Diagnostics, catalog number: 10 901 393 001 )
- Pre-stained Protein Ladder (broad range) (New England Biolabs, catalog number: P7710S )
- Goat anti-UPF1 Antibody (G-α-RENT1) (Bethyl Laboratories, catalog number: A300-038A )
- Goat anti-rabbit IgG (polyclonal) (Bio-Rad Laboratories, catalog number: 172-1053 )
- Rabbit anti-actin (polyclonal) (Sigma-Aldrich, catalog number: A5050 )
- AffiniPure Goat Anti-mouse IgM (µ chain specific) (Jackson ImmunoResearch Laboratories, catalog number: 115-005-020 )
- IRDye 800CW Donkey anti-Rabbit (LI-COR, catalog number: 926-32213 )
- IRDye 800CW Donkey anti-Goat (LI-COR, catalog number: 926-32214 )
- Dynabeads® Protein G (Life Technologies, catalog number: 10004D )
- Wizard® SV Gel and PCR Clean-Up System (Promega Corporation, catalog number: A9282 )
- Affinity Script Multi-Temp Reverse Transcriptase (Agilent, catalog number: 600105 )
- Random hexamer primers (Microsynth AG)
- Brilliant III Ultra-Fast qPCR Master mix (Agilent, catalog number: 600880 )
- Chloroform
- Isopropanol
- DMEM-/- (see Recipes)
- DMEM+/+ (see Recipes)
- Phosphate-bufferd saline (PBS) (pH 7.4) (see Recipes)
- Hypotonic gentle lysis buffer (pH 7.5) (RNase-free) (see Recipes)
- Wash buffer (pH 7.5) (RNase-free) (see Recipes)
- Net-2 buffer (pH 7.5) (RNase-free) (see Recipes)
- Hybridization buffer (pH 7.5) (see Recipes)
- 2x SDS loading buffer (pH 6.8) (see Recipes)
- TRI-reagent (see Recipes)
- Tris buffered saline (pH 7.6) (TBS) (see Recipes)
- TBS-Tween milk (see Recipes)
- Bjerrum transfer buffer (see Recipes)
- DEPC treated water/buffer (see Recipes)
- Turbo DNase mix (see Recipes)
Equipment
- Pure water system: PURELAB Priama (Prima 7) and PURELABULTRA (Ultra Genetic) (ELGA LabWater)
- CO2 incubator (BINDER GmbH, model: 9140-0047 )
- Clear-viewTM Snap-Cap microtubes (1.5 ml, natural, low retention) (Sigma-Aldrich, catalog number: T4816-250EA )
- Multiply®-Pro 0.2Ml Biosphere® (Sarstedt AG, catalog number: 72.727 )
- Filter Tips (10 µl, 20 µl and 200 µl) (Axon Lab AG, catalog numbers: AL60X10 , AL60X20 , AL60X200 )
- Filter Tips (1,250 µl) (Greiner Bio-One GmbH, catalog number: 7.750.261 )
- CountessTM automated cell counter (Life Technologies, model: C10227 )
- CountessTM cell counting chamber slides (Life Technologies, catalog number: C10283 )
- 15 cm tissue culture dishes (TPP Techno Plastic Products, catalog number: 93150 )
- GP Millipore express® PLUS Membrane (0.22 µm) (500 ml Funnel, 45 mm Neck Size) (EMD Millipore, catalog number: SCGPT05RE )
- NanoDrop 2000 (Thermo Fisher Scientific)
- Heat block, Thermomixer® compact (Eppendorf)
- DynaMagTM-2 magnet (Life Technologies, catalog number: 12321D )
- Eppendorf centrifuge 5415R with rotor F45-24-11 (Eppendorf, catalog numbers: 022621459 and 022636502 )
- Lab cycler gradient equipped with Thermoblock 96 (SensoQuest GmbH, models: 011-101 and 012-103 )
- Blotting paper (ALBET Lab Science, catalog number: BP 002 46579 )
- Corbett Rotor-Gene® 6000 (QIAGEN)
- Corbett CAS-1200 (QIAGEN)
- SE260 Mighty Small II Deluxe Mini vertical electrophoresis unit (Hoefer, model: SE260-10A-1.5 )
- Optitran BA-S 85 reinforced nitrocellulose membrane (GE Healthcare, Whatman, catalog number: 10 439 196 )
- Semi-dry transfer unit with built-in power supply TE77XP (Hoefer, model: TE77XP )
- Odyssey® infrared imaging system (LI-COR)
Procedure
- Cloning of the p-β SL mini µ WT uA1 plasmid
To generate the p-β SL mini µ WT uA1 plasmid, a double-stranded oligonucleotide encoding a stable stem-loop structure is introduced into the KpnI site of p-β mini µ WT uA1 plasmid (Yepiskoposyan et al., 2011).- Open the p-β mini µ WT uA1 vector by KpnI restriction digestion
- Digest 4.2 µg p-β mini µ WT uA1 plasmid with 36 U Kpn I in a total volume of 40 µl according to the manufaturer’s protocol at 37 °C for 1 h.
- Add 5 µl 10x CIP Buffer and 5 µl CIP (1 U/µl) and incubate at 37 °C for 30 min to dephosphorylate the open vector.
- Run the digested vector on 1% agarose gel, excise the DNA band and isolated the vector using the Wizard SV Gel PCR Clean-Up System.
- Digest 4.2 µg p-β mini µ WT uA1 plasmid with 36 U Kpn I in a total volume of 40 µl according to the manufaturer’s protocol at 37 °C for 1 h.
- Anneal SL Oligo 1 and SL Oligo 2
- Mix 1 µl SL oligo 1 (100 µM), 1 µl SL oligo 2 (100 µM) and 48 µl hybridization buffer.
- Heat the hybridization mix to 90 °C and slowly cool down to 40 °C (in a heat block).
- To phosphorylate the double stranded oligonucleotide, combine 2 µl of the hybridization mix, 24 µl MQ-water, 3 µl 10x T4 DNA ligase buffer, 1 µl T4 PNK and incubate at 37 °C for 30 min.
- Inactivate the kinase at 70 °C for 15 min.
- Mix 1 µl SL oligo 1 (100 µM), 1 µl SL oligo 2 (100 µM) and 48 µl hybridization buffer.
- Ligate 50 ng of the open p-β mini µ WT uA1 vector with 2 µl of the double-stranded oligonucleotide in 1x T4 DNA ligase buffer with 1 µl of T4 DNA ligase (400 U/µl) in a total volume of 15 µl at room temperature for 2.5 h.
- Transform the 5 µl of ligation mix into 100 µl XL10-Gold ultracompetent cells.
- The presence of the sequence encoding the stem-loop preceding the mini µ open reading frame is verified by sequencing.
- Open the p-β mini µ WT uA1 vector by KpnI restriction digestion
- Transfect hela cells either with p-β SL mini µ WT uA1 or p-β mini µ WT uA1 plasmid
- Hela cells are cultivated in DMEM supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin and 10% FCS (DMEM+/+: DMEM with antibiotics and FCS).
- (Day 1) Seed 3 x 106 hela cells into each of two 15 cm diameter dishes to have 60% confluent cells the next day.
- (Day 2) Cells were transfected either with 5 µg p-β mini µ WT uA1 or 5 µg p-β SL mini µ WT uA1 plasmid using 40 µl DreamFectTM according the manufacturer’s protocol.
- (Day 3) Split the cells from one plate into two 15 cm diameter dishes.
- (Day 4) Harvest the cells.
- Wash the cells with 25 ml PBS.
- To detach the cells they are incubated with 4 ml trypsin-EDTA (T/E) at 37 °C for 10 min.
- Add 9 ml DMEM+/+ and resuspend the cells.
- Count the cells using the Countess™ automated cell counter according to the manufacturer’s protocol.
- Wash the cells with 25 ml PBS.
- Hela cells are cultivated in DMEM supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin and 10% FCS (DMEM+/+: DMEM with antibiotics and FCS).
- Lyse the cells
- 1.2 x 107 cells are lysed on ice in 1.2 ml hypotonic gentle lysis buffer supplemented with 40 U/ml RiboLock RI RNase inhibitor for 20 min.
- Clear the lysate by centrifugation in a microcentrifuge (16,000 x g, 4 °C, 15 min).
- 1.2 x 107 cells are lysed on ice in 1.2 ml hypotonic gentle lysis buffer supplemented with 40 U/ml RiboLock RI RNase inhibitor for 20 min.
- Take protein and RNA input samples
- Protein input samples are prepared by mixture of 50 µl lysate with 50 µl SDS loading buffer, cooked at 90 °C for 5 min and stored at -20 °C.
- Prior to the extraction of input RNA, 100 µl cell lysates are supplemented with 2.5 µl RiboLock RI RNase inhibitor and treated with 20 U/ml TURBO DNase at 37 °C for 5 min. Subsequently, the RNA was extracted with TRI reagent according the following TrIzol protocol.
- Mix 100 µl TURBO DNase treated cell lysate with 900 µl TRI reagent, vortex.
- Incubate at room temperature for 5 min.
- Add 180 ml chloroform to each sample and vortex for 15 s.
- Incubate at RT for 5 min.
- Centrifuge the samples (12,000 x g, 4 °C, 15 min), lift 350 µl of the aqueous phase and transfer into a fresh test tube.
- Add 2 µl glycogen, 450 µl isopropanol, vortex and incubate at -20 °C for 30 min.
- Precipitate the RNA by centrifugation (12,000 x g, 4 °C, 10 min).
- Discard the supernatant and wash the pellet with 1 ml 70% EtOH.
- Spin the samples (12,000 x g, 4 °C, 10 min), discard the supernatant and air dry the pellet.
- Resolve the RNA in 60 µl MQ-water.
- Measure the RNA concentration on the NanoDrop 2000. A ratio of OD260/OD230 < 2 is indicative for phenol contamination and demands for an additional EtOH precipitation of the samples.
- Mix 100 µl TURBO DNase treated cell lysate with 900 µl TRI reagent, vortex.
- Protein input samples are prepared by mixture of 50 µl lysate with 50 µl SDS loading buffer, cooked at 90 °C for 5 min and stored at -20 °C.
- UPF1 RNA immunopurification (RIP)
- Adapt the NaCl concentration to 150 mM by the addition of 52.5 µl 3 M NaCl to the remaining 1,050 µl cell lysates expressing either the mini µ or SL mini µ constructs.
- 500 µl of each lysate is incubated head-over-tail at 4 °C for 90 min with 3.7 µg goat anti-RENT1 and goat anti-rabbit IgG antibody, respectively (for RIP incubation procedure see Table 1).
- Preparation of the Dynabeads® protein G beads.
- 128 µl Dynabeads® protein G beads are washed with 1 ml wash buffer.
- Beads are washed with 1 ml hypotonic gentle lysis buffer.
- Beads are equilibrated in hypotonic gentle lysis buffer supplemented with 1% (w/v) BSA and 0.1% yeast tRNA at 4 °C for 1 h (to minimize unspecific binding).
- Wash the beads twice with 1 ml hypotonic gentle lysis buffer.
- 128 µl Dynabeads® protein G beads are washed with 1 ml wash buffer.
- The Dynabeads® are equally distributed on the four RIP samples and incubated head over tail at 4 °C for 90 min (Table 1).
Table 1. UPF1 RIP incubation schema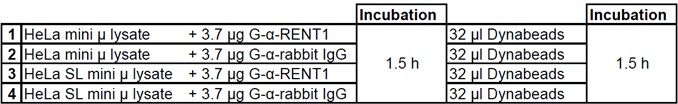
- Precipitates were washed six times with 1 ml Net-2 buffer.
- During the last wash step the beads are separated.
- One-third of the beads are incubated with 40 µl 2x SDS loading buffer at 90 °C for 5 min in order to elute the protein for subsequent western blot analysis.
- Two-thirds of the beads are dissolved in 50 µl Turbo DNase mix and DNase treated at 37 °C for 10 min. To isolate the RNA 900 µl TRI reagent is added to the samples and treated according to the Trizol protocol (see 4b). The RNA is dissolved in 34.5 µl MQ-water, reverse transcribed and analyzed by RT-qPCR.
- One-third of the beads are incubated with 40 µl 2x SDS loading buffer at 90 °C for 5 min in order to elute the protein for subsequent western blot analysis.
- Adapt the NaCl concentration to 150 mM by the addition of 52.5 µl 3 M NaCl to the remaining 1,050 µl cell lysates expressing either the mini µ or SL mini µ constructs.
- Quantitative real-time reverse-transcription PCR
- The entire recovered RNA of the UPF1 RIPs, or 1 µg of the input RNA samples are reverse transcribed in a total volume of 50 µl containing 1x StrataScript RT buffer, 0.1 mM DTT, 0.4 mM dNTPs, 300 ng random hexamer primers and 1 µl of StrataScript Multi-Temp reverse transcriptase according to the manufacturere’s protocol.
- Twelve microliters of the reverse transcription reactions were amplified with Brilliant III Ultra-Fast qPCR Master mix in the Corbett Rotor-Gene 6000 (Pipetting was done using the Corbett CAS-1200 robot.). The primers and TaqMan probes to measure SMG5 and mini µ mRNA levels are described elsewhere (Yepiskoposyan et al., 2011) (see also Table 2). The levels of SMG5 mRNA, which efficiently associate with UPF1 (Yepiskoposyan et al., 2011) are measured to normalize for variable RIP efficiencies in different samples.
Table 2. TaqMan probes
- The entire recovered RNA of the UPF1 RIPs, or 1 µg of the input RNA samples are reverse transcribed in a total volume of 50 µl containing 1x StrataScript RT buffer, 0.1 mM DTT, 0.4 mM dNTPs, 300 ng random hexamer primers and 1 µl of StrataScript Multi-Temp reverse transcriptase according to the manufacturere’s protocol.
- Western blotting
- For protein analysis, 2 x 105 cells and one-third of the RIP samples are separated on a 10% SDS-PAGE using the Mighty Small II running chamber SE260. The proteins are transferred to Optitran BA-S 85 reinforced nitrocellulose membrane using a semi dry blotter and Bjerrum transfer buffer.
- After blocking in TBS-Tween milk, the membrane is probed in the blocking buffer with 1:3,000-diluted polyclonal goat anti-RENT1 antibody to assess the specificity of the UPF1 RIP experiment, 1:500-diluted AffiniPure goat anti-mouse IgM, µ chain-specific antibody to compare mini µ expression levels either from p-β SL mini µ WT uA1 or p-β mini µ WT uA1 plasmid or 1:5,000-diluted polyclonal rabbit anti-actin antibody to be used as a loading control.
- The membrane is probed with 1:10,000-diluted donkey anti-rabbit IRDye 800CW or donkey anti-goat IRDye 800CW antibodies.
- The membrane is scanned on an Odyssey infrared imager.
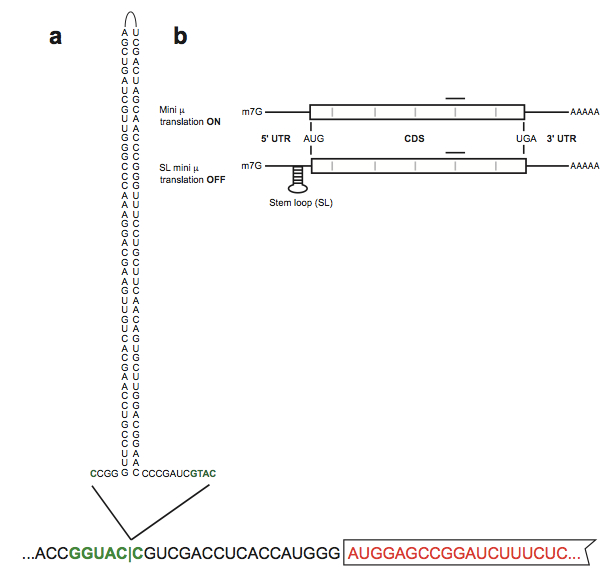
Figure 1. Stem loop-induced stranscript specific translation inhibition. a. Representation of the stem loop structure introduced in the KpnI site (green palindromic sequence) preciding the mini µ open reading frame (red boxed sequence) of the Ig-µ reporter transcript mini µ. b. Schematic representation of mini µ and SL mini µ reporter transcripts. The translation-inhibing stable stem loop in the 5' UTR of SL mini µ is shown. The CDS is represented as a white box flanked by the 5' and 3' UTRs. Gray bars, exon-exon junctions; black line, position of the TaqMan assay used for real-time qPCR with reverse transcription (RT-qPCR) (adapted from Reference 1) - The membrane is probed with 1:10,000-diluted donkey anti-rabbit IRDye 800CW or donkey anti-goat IRDye 800CW antibodies.
Recipes
Note: All buffers used in the RIP protocol have to be sterile and RNase-free. Therefore, all buffers are prepared using MQ-water and if possible DEPC treated. Duran bottles, glassware and spatulas are baked at 180 °C for 2 h. Polycarbonate or polystyrene materials (e.g. magnetic stirrers…) are soaked in 3% hydrogen peroxide or 2 M NaOH for 10 min and extensively rinsed with DEPC-treated water. If a buffer cannot be DEPC treated (e.g. Tris buffers) prepare the buffer in DEPC or MQ-water and filter it with a 0.22 µm filter.
- DMEM-/- (per 900 ml)
12 g DMEM/F12 powder
2.48 g NaHCO3
Make up to 900 ml with ddH2O
Adjust pH to 7.2 with 32% HCl or 10 M NaOH
Sterilize per filtration with bottle top filter (0.22 µm) - DMEM+/+ (per 500 ml)
Supply 450 ml DMEM-/- with 50 ml FCS and 5 ml P/S - Phosphate-bufferd saline (PBS)
137 mM NaCl
10 mM Na2HPO4
2.7 mM KCl
2 mM KH2PO4
Adjust pH to 7.4 with HCl - Hypotonic gentle lysis buffer (RNase-free)
10 mM Tris-HCl (pH 7.5)
10 mM NaCl
2 mM EDTA
0.5% (v/v) Triton X-100
Prior to usage supplement with 1x HaltTM protease inhibitor cocktail - Wash buffer (RNase-free)
50 mM Tris-HCl (pH 7.5)
150 mM NaCl
0.05% (v/v) NP-40
Prior to usage supplement with 1x HaltTM protease inhibitor cocktail - Net-2 buffer (RNase-free)
150 mM NaCl
50 mM Tris-HCl (pH 7.5)
0.1% (v/v) Triton-X-100
Prior to usage supplement with 1x HaltTM protease inhibitor cocktail - Hybridization buffer
100 mM KOAc
30 mM HEPES-KOH (pH 7.5)
2 mM MgOAc - 2x SDS loading buffer
200 mM DTT
120 mM Tris-HCl (pH 6.8)
0.44% (w/v) SDS
20% (v/v) glycerol
0.25% (w/v) bromophenol blue - TRI-reagent
800 mM guanidine thiocyanate
400 mM ammonium thiocyanate
100 mM sodium acetate
38% (v/v) phenol
5% (v/v) glycerol
0.1% (w/v) 8-quinolinol (pH 5.0) - Tris buffered saline (TBS)
137 mM NaCl
20 mM Tris-HCl (pH 7.6) - TBS-Tween milk
5% (w/v) milk powder, fat-free
0.1% (v/v) Tween-20
Make up to 250 ml using TBST - Bjerrum transfer buffer
48 mM Tris base
39 mM glycin
0.1% (w/v) SDS
20 (v/v) MeOH - DEPC treated water/buffer
0.1% (v/v) DEPC
Stir over night at 4 °C
Autoclave twice in order to inactivate DEPC - Turbo DNase mix
1x Turbo DNase buffer
0.1 U/µl Turbo DNase
1 U/µl RiboLock RNase inhibitor
Acknowledgments
We thank L. Maquat (University of Rochester, Rochester, New York, USA) for providing their protocol for RIP coupled with RNase-H cleavage, and S. Rufener (Department of Chemistry and Biochemistry, University of Bern, Bern, Switzerland) for help with the RIPs. This work was supported by grants to Oliver Mühlemann from the European Research Council (StG 207419), the Swiss National Science Foundation (31003A-127614 and 31003A-143717) and the canton of Bern.
References
- Yepiskoposyan, H., Aeschimann, F., Nilsson, D., Okoniewski, M. and Mühlemann, O. (2011). Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 17(12): 2108-2118.
- Zünd, D., Gruber, A. R., Zavolan, M. and Mühlemann, O. (2013). Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3' UTRs. Nat Struct Mol Biol 20(8): 936-943.
- For protein analysis, 2 x 105 cells and one-third of the RIP samples are separated on a 10% SDS-PAGE using the Mighty Small II running chamber SE260. The proteins are transferred to Optitran BA-S 85 reinforced nitrocellulose membrane using a semi dry blotter and Bjerrum transfer buffer.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zünd, D. and Mühlemann, O. (2014). UPF1 RNA Immunoprecipitation from Mini-μ Construct–expressing Cells. Bio-protocol 4(7): e1086. DOI: 10.21769/BioProtoc.1086.
Category
Biochemistry > RNA > RNA-protein interaction
Molecular Biology > RNA > RNA detection
Biochemistry > Protein > Immunodetection > Immunoprecipitation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









