- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
RNA Isolation and Northern Blot Analysis
Published: Vol 4, Iss 6, Mar 20, 2014 DOI: 10.21769/BioProtoc.1077 Views: 33864
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Purification of Bacterial RNA from Infected Macrophages
Lior Lobel [...] Anat A. Herskovits
Nov 20, 2015 11198 Views

Single Genome Sequencing of Expressed and Proviral HIV-1 Envelope Glycoprotein 120 (gp120) and nef Genes
David J. Nolan [...] Michael S. McGrath
Jun 20, 2017 10043 Views
Abstract
The northern blot is a technique used in molecular biology research to study gene expression by detection of RNA in a sample. With northern blotting it is possible to observe particular gene expression levels during differentiation, morphogenesis, as well as abnormal or diseased conditions. Here, we examine ATF3, ATF4, and GADD153 gene expression profiles by northern blot in Vero cells and H1299 cells after IBV infection. RNA was extracted in IBV (infectious bronchitis virus) infected cells and electrophoresis was used to separate the RNA sample. RNA was transferred from the electrophoresis gel to the blotting membrane by capillary transfer. Specific mRNA was detected with hybridization probes complementary to part of target sequence. The probes were prepared by RT-PCR and labeled by digoxigenin (DIG) using DIG labeling kit.
Materials and Reagents
- Vero cells (kidney epithelial cells extracted from an African green monkey) (ATCC, catalog number: CCL-81 TM)
- H1299 cells (human lung carcinoma cell line) (ATCC, catalog number: CRL-5803 TM)
- The egg-adapted Beaudette strain of IBV (ATCC, catalog number: VR-22 )
- Dulbecco modified Eagle medium (DMEM) (Life Technologies, Gibco®, catalog number: 11960-044 )
Note: It contains more vitamins and more glucose, as well as iron and is suitable for most types of cells. - Roswell Park Memorial Institute medium (RPMI) 1640 (Life Technologies, Gibco®, catalog number: 21870-076 )
Note: This medium contains a great deal of phosphate and is formulated for use in a 5% carbon dioxide atmosphere. - Trypsin/EDTA (Life Technologies, Gibco®, catalog number: 25200-072 )
- TRIzol reagent (Life Technologies, Gibco®, catalog number: 15596-018 )
- Chloroform (Thermo Fisher Scientific, catalog number: C/4960/17 )
- Isopropanol (Thermo Fisher Scientific, catalog number: P/7507/17 )
- Ethanol (Merck KGaA, catalog number: 1.00983.2500 )
- RNase free water
- Reverse transcriptase AMV (Roche Diagnostics, catalog number: 10109118001 )
- Oligo (dT) (1st Base Biochemicals)
- RNasin® ribonuclease inhibitor (Promega Corporation, catalog number: N2511 )
- Primers (1st Base Biochemicals)
- DIG labeling kit (Roche, catalog number: 11175025910 )
- RNA loading buffer (New England Biolabs, catalog number: B0363S )
- Agarose (1st Base Biochemicals, catalog number: BIO-100-500G )
- Formaldehyde (Thermo Fisher Scientific, catalog number: F75P1GAL ))
- Ethidium bromide (Bio-Rad Laboratories, catalog number: 1610433 )
- HybondTM-N+ membrane (Amersham Biosciences, catalog number: RPN303B )
- DIG Wash and Block Buffer Set (Roche Diagnostics, catalog number: 11585762001 )
- DIG easy Hyb (Roche Diagnostics, catalog number: 11603558001 )
- Anti-digoxigenin-AP fab fragments (Roche Diagnostics, catalog number: 11093274910 )
- CDP-Star (Roche Diagnostics, catalog number: 12041677001 )
- Amersham hyperfilm ECL (Amersham Biosciences, catalog number: 28906837 )
- 70% RNase-free ethanol
- Tris(hydroxymethyl)aminomethane (Tris base) (Promega Corporation, catalog number: H5135 )
- Acetic acid (Glacial) (Merck KGaA, catalog number: 1.00063.2500 )
- 3-(4-morpholino) propane sulfonic acid (MOPS) (Thermo Fisher Scientific, catalog number: BP308-500 )
- Sodium acetate.3H2O (Thermo Fisher Scientific, catalog number: S207-10 )
- Sodium Citrate (Thermo Fisher Scientific, catalog number: S25545 )
- 10x TAE Electrophoresis Buffer (1 L) (see Recipes)
- 10x MOPS buffer (1 L) (see Recipes)
- 1x MOPS buffer (1 L) (see Recipes)
- 1.3% Formaldehyde Agarose gel (see Recipes)
- 20x SSC buffer (1 L) (see Recipes)
- 2x SSC, 0.1% SDS (1 L) (see Recipes)
- 0.1x SSC, 0.1% SDS (see Recipes)
Equipment
- 100 mm cell culture dishes (Corning, catalog number: 430167 )
- 0.2 ml thin-wall Gene-Amp PCR tube (Corning, Axygen®, catalog number: PCR-02-C )
- FormaTM Steri-CycleTM CO2 Incubators (Thermo Fisher Scientific, catalog number: 201370 )
- OLYMPUS CKX31 microscope
- Eppendorf centrifuge 5415R
- NanoDrop (Thermo Fisher Scientific, model: ND-1000 spectrophotometer )
- Power Pac and electrophoresis tank (Bio-Rad Laboratories)
- Tray
- Glass plate
- Tissue paper
- CL-1000, ultraviolet crosslinker (UVP)
- Hybaid Maxi 14 Hybridization Oven (Thermo Fisher Scientific)
- Hybridization tubes
- Kodak Biomax cassette (Eastman Kodak Company)
- Kodak X-OMAT 2000 processor (Eastman Kodak Company)
Procedure
- RNA extraction
- Cells were seeded in 100-mm-diameter dishes and infected with either 2 PFU of live IBV per cell or the same amount of UV-inactivated IBV (UV-IBV) at 37 °C. Excess virus in the medium was removed by replacing with fresh medium at 1 h post-infection.
- The IBV-infected cells were incubated at 37 °C in 5% CO2.
- At the indicated time points (0, 2, 4, 8, 12, 16, 20, 24, 28 h post-infection), cells were rinsed with 10 ml Phosphate Buffered Saline (PBS) buffer once and lysed in 1 ml TRIzol for 5 min at room temperature.
- Cell lysates were transfer into eppendorf tubes and one-fifth (volume/volume) of chloroform was added to each tube.
- Shake tubes vigorously by hand for 15 sec and incubated for 3 min at room temperature, then centrifuged at 12,000 x g for 15 min at 4 °C.
- The upper aqueous phase was transfer into a new tube and mixed with 1:1 (volume/volume) of 100% isopropanol, and then incubated for 10 min at room temperature.
- RNA was precipitated by centrifugation at 12,000 x g for 10 min at 4 °C.
- RNA pellet was washed with 1 ml 70% RNase-free ethanol once and spin down by 7,500 x g for 5 min.
- The RNA pellets are air-dried and dissolved in 100 µl RNase-free H2O by incubating at 65 °C for 15 min.
- RNA concentration and purity were determined by NanoDrop.
- The RNAs were stored at -80 °C for further use.
- Cells were seeded in 100-mm-diameter dishes and infected with either 2 PFU of live IBV per cell or the same amount of UV-inactivated IBV (UV-IBV) at 37 °C. Excess virus in the medium was removed by replacing with fresh medium at 1 h post-infection.
- Probe preparation
- Northern blot probes were obtained by RT-PCR and labeled by digoxigenin (DIG) using DIG labeling kit described as follow steps.
- 2 µg of total RNA is added to 2 µl of 10 pmoles of an oligo (dT) in a sterile 0.2 ml thin-wall Gene-Amp PCR tube of a final volume of 10.5 µl.
- After denaturation at 65 °C for 10 min, the tubes are cooled on ice immediately.
- The denatured RNA-primer mixture is then added to a final volume of 20 µl reaction mixture containing 10 mM of dNTPs, 20 units of Rnasinâ ribonuclease inhibitor, 1x Expandä reverse transcriptase buffer and 50 units of reverse transcriptase.
- The first strand cDNA is synthesized at 43 °C for 1 h, and reaction can be terminated by heating at 65 °C for 10 min (optional).
- Amplification of cDNA was achieved by polymerase chain reaction (PCR) in a 25 or 50 µl reactions containing of appropriate primer pairs and PFU polymerase using the DIG labeling kit according to the manufacturer’s manual.
- Primers used for human ATF4 were 5’-CCGTCCCAAACCTTACGATC-3’ (forward) and 5’-ACTATCCTCAACTAGGGGAC-3’ (reverse). Primers used for human ATF3 were 5’-GGTTAGGACTCTCCACTCAA-3 (forward) and 5’-AGACAGTAGCCAGCGTCCTT-3’ (reverse). Primers used for human GADD153 were 5'-GATTCCAGTCAGAGCTCCCT3' (forward) and 5'-GTAGTGTGGCCCAAGTGGGG-3' (reverse). Prepare a 10x concentration solution of each respective PCR primer.
- Add the following reagents in a 0.2 ml reaction tube on ice, in the following order: ddH2O 32.25 µl, PCR buffer 5 µl, PCR DIG labeling mix 5 µl, forward primer 5 µl, reverse primer 5 µl, enzyme mix 0.75 µl, template cDNA 2 µl, final volume 50 µl. Vortex the mixture and centrifuge briefly.
- Place the sample in a thermal block cycler and perform PCR in following condition: initial denature at 95 °C for 2 min, denature at 95 °C for 10 sec, anneal at 60 °C for 30 sec, and elongate at 72 °C for 2 min, repeat denaturation, annealing, and elongation for 30 cycles, finally elongate at 72 °C for 7 min.
- Run a portion of each PCR reaction on an agarose mini gel and then stain the gel with ethidium bromide and examine the PCR products under UV.
- Northern blot probes were obtained by RT-PCR and labeled by digoxigenin (DIG) using DIG labeling kit described as follow steps.
- Northern blot
- To analyze RNA expression by Northern blot, 30 µg of RNA from each sample preparation was mixed with RNA loading buffer and load on wells in 1.3% agarose formaldehyde gel (see Recipes).
- Run the gel with 3-4 V/cm in RNase free gel boxes for 4 h until the RNAs are well separated.
- Stain the gel briefly in 0.25 – 0.5 µg/ml ethidium bromide and examine the gel under UV light.
- Rinse gels for 2 x 15 min in 20x SSC and RNA on the gel were transferred onto a HybondTM-N+ membrane by capillary transfer with 20x SSC overnight at room temperature.
- Fix the RNA to the membrane by UV-crosslinking. The energy used is 20,000 µJoules/cm2 at 245 nm.
- After the UV-crosslinking, rinse the membranes briefly in ddH2O and allow to air-dry.
- Prehybridize membranes with DIG easy Hyb for 30 min with gentle agitation at 68 °C.
- Denature DIG-labeled RNA probes by boiling for 5 min and rapidly cooling in ice, and add the denatured probes (25 ng/ml) to 10 ml prewarmed DIG Easy Hyb.
- 10 ml probe/hybridization mixtures were added to membranes and incubated for 6 h at 68 °C with gentle agitation.
- After hybridization, membranes were washed with 2x SSC, 0.1% SDS for 2 x 5 min at 25 °C under constant agitation, and then washed with 0.1x SSC, 0.1% SDS for 2 x 15 min at 68 °C under constant agitation.
- The membranes were then rinsed briefly (5 min) in washing buffer and was blocked in blocking buffer for 30 min.
- After blocking, membranes were incubated with DIG antibody (Dilute anti-DIG-AP 1:10,000 in blocking buffer) for 30 min, washed 2 x 15 min in washing buffer and equilibrated 3 min in detection buffer.
- The signal was detected with CDP-Star according to the manufacturer’s instructions (see Figures 1-3).
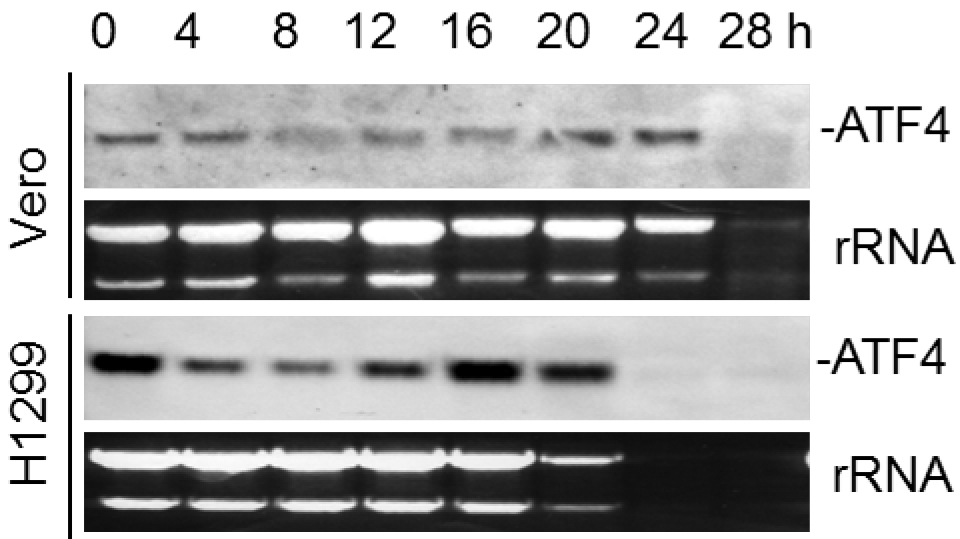
Figure 1. Northern blot analysis of ATF4 mRNA in IBV-infected cells. Vero and H1299 cells were infected with IBV (MOI~1) and harvested at indicated time points. Total RNA was isolated and subjected to Northern blot using an ATF4 probe. Ethidium bromide staining of 28S rRNA and 18S rRNA is shown as a loading control. Band intensities of ATF4 were determined and normalized to rRNA.
Figure 2. Northern blot analysis of ATF3 mRNA in IBV-infected cells. Vero and H1299 cells were infected as in Figure 1. RNA extraction and Northern blot was performed as in Figure 1 using ATF3 probe.
Figure 3. Northern blot analysis of GADD153 at the mRNA level in IBV-infected cells. Vero and H1299 cells were infected with IBV (MOI~1) or incubated with UV-IBV and harvested at indicated time points. RNA extraction and Northern blot was performed as in Figure 1 using GADD153 probe.
- To analyze RNA expression by Northern blot, 30 µg of RNA from each sample preparation was mixed with RNA loading buffer and load on wells in 1.3% agarose formaldehyde gel (see Recipes).
Recipes
- 10x TAE Electrophoresis Buffer (1 L)
48.4 g Tris(hydroxymethyl)aminomethane (Tris base)
11.4 ml 17.4 M glacial acetic acid
3.7 g EDTA, disodium salt
ddH2O - 10x MOPS buffer (1 L)
83.7 g 3-(N-morpholino) propanesulfonic acid (MOPS)
13.61 g Sodium acetate.3H2O
3.7 g EDTA
ddH2O - 1x MOPS buffer (1 L)
100 ml 10x MOPS buffer
20 ml 37%-formaldehyde
880 ml ddH2O - 1.3% Formaldehyde Agarose gel
1.3 g agarose
10 ml 10x Formaldehyde Agarose gel buffer
Add RNase-free water to 100 ml
Heat the mixture to melt agarose
Cool to 65°C in a water bath.
Add 1.8 ml of 37% (12.3 M) formaldehyde (toxic) and 1 µl of a 10 mg/ml ethidium Bromide stock solution
Mix thoroughly and pour onto gel support
Prior to running the gel, equilibrate in 1x Formaldehyde Agarose gel running buffer for at least 30 min - 20x SSC buffer (1 L)
175.3 g of NaCl
88.2 g of Sodium Citrate
ddH2O
Adjust the pH to 7.0 with a few drops of 14 N solution of HCl
Sterilized by autoclaving - 2x SSC, 0.1% SDS (1 L)
100 ml 20x SSC buffer
10 ml 10%SDS
890 ml ddH2O - 0.1x SSC, 0.1% SDS
10 ml 20x SSC buffer
10 ml 10%SDS
980 ml ddH2O
Acknowledgments
This protocol was adapted from the previous publication Liao et al. (2013). This work was partially supported by a Competitive Research Programme (CRP) grant (R-154-000-529-281) from the National Research Foundation, Singapore.
References
- Liao, Y., Fung, T. S., Huang, M., Fang, S. G., Zhong, Y. and Liu, D. X. (2013). Upregulation of CHOP/GADD153 during coronavirus infectious bronchitis virus infection modulates apoptosis by restricting activation of the extracellular signal-regulated kinase pathway. J Virol 87(14): 8124-8134.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Liao, Y., Fung, T. S., Huang, M., Fang, S., Zhong, Y. and Liu, D. (2014). RNA Isolation and Northern Blot Analysis. Bio-protocol 4(6): e1077. DOI: 10.21769/BioProtoc.1077.
Category
Microbiology > Microbial genetics > RNA > RNA extraction
Molecular Biology > RNA > RNA extraction
Molecular Biology > RNA > RNA detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










