- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Regulatory T cells Differentiation From Naïve T Cells
Published: Vol 4, Iss 6, Mar 20, 2014 DOI: 10.21769/BioProtoc.1075 Views: 24283
Reviewed by: Lin FangOmar AkilFanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Differentiation of THP1 Cells into Macrophages for Transwell Co-culture Assay with Melanoma Cells
Michael Peter Smith [...] Claudia Wellbrock
Nov 5, 2015 50339 Views

3D Co-culture System of Tumor-associated Macrophages and Ovarian Cancer Cells
Lingli Long [...] Wang Min
Apr 20, 2018 12145 Views

Accurate Identification of Cell Cycle Stages in RPE1 Cells Using the ImmunoCellCycle-ID Method
Syon Reddy [...] Aussie Suzuki
Aug 5, 2025 1876 Views
Abstract
In the past years, a subset of regulatory T cells (Tregs) expressing CD4, CD25 and the transcription factor FoxP3 has gained considerable attention as key regulators of T-cell tolerance and homeostasis (Sakaguchi, 2004). This population of T cells is specifically engaged in the maintenance of immune self-tolerance and the control of aberrant immune responses to foreign antigens. Remarkably, regulatory T cells have been implicated in tumor cell evasion of immune responses (Curiel et al., 2004; Zou, 2006) by suppressing T cell mediated antitumor immunity. The study of the signals that promote the differentiation of this suppressive population in the tumor microenvironment has become a central issue. Here we described a detailed method to in vitro differentiate Tregs using tumor cells conditioned media from mouse naïve T cells and to identify them based on their specifics markers (Dalotto-Moreno et al., 2013).
Keywords: Regulatory T cellMaterials and Reagents
- Splenocyte suspension
- Eight- to twelve-week old Balb/c mice strain
- RPMI 1640 (Life Technologies, Gibco®, catalog number: 22400-089 )
- Phosphate buffer saline (PBS) (see Recipes)
- Sterile red blood lysis buffer (ACK buffer) (see Recipes)
- Eight- to twelve-week old Balb/c mice strain
- Cell lines
- 4T1 cell line (ATCC)
4T1 is a highly metastatic stage IV murine breast cancer cell line that lacks estrogen and progesterone nuclear receptors and that spontaneously metastasizes to lung, brain and bone.
- RPMI 1640
- Heat-inactivated fetal bovine serum (FBS) (Life Technologies, Gibco®, catalog number: 10438-026 )
- 100x Antibiotic-antimycotic (Life Technologies, InvitrogenTM, catalog number: 15240062 )
- 4T1 cell line (ATCC)
- Determination and purification of CD4+ Treg and naïve T cells
- Allophycocyanin (APC)-conjugated anti-CD4 antibody (clone GK1.5) (eBioscience, catalog number: 17-0041 )
- Alexa Fluor 488-conjugated anti-CD25 antibody (clone PC61.5) (eBioscience, catalog number: 53-0251 )
- Phycoerythrin (PE) -conjugated anti-CD62L antibody (clone MEL-14) (eBioscience, catalog number: 12-0621 )
- PE-conjugated anti-Foxp3 antibody (clone FJK-16s) (eBioscience, catalog number: 12-5773 )
- Fix/Perm buffer (eBioscience, catalog number: 00-5123 , 00-5223 )
- 10x Permeabilization Buffer (eBioscience, catalog number: 00-8333 )
- Dynal® Mouse CD4 Cell Negative Isolation Kit (Life Technologies, InvitrogenTM, catalog number: 114-15D )
- Heat-inactivated fetal bovine serum (FBS) (Life Technologies, Gibco®, catalog number: 10438-026)
- FACS buffer (see Recipes)
- Sorted cells collection medium (see Recipes)
- Allophycocyanin (APC)-conjugated anti-CD4 antibody (clone GK1.5) (eBioscience, catalog number: 17-0041 )
- Differentiation of Treg in vitro
- NA/LE Hamster anti-mouse CD3ε monoclonal antibody (clone 145-2C11) (BD, catalog number: 553057 )
- NA/LE Hamster anti-mouse CD28 monoclonal antibody (clone 37.51) (BD, catalog number: 553294 )
- Antibiotic-antimycotic (Life Technologies, InvitrogenTM, catalog number: 15240062)
- RPMI 1640 supplemented with 50 µM β-mercaptoethanol and antibiotic-antimycotic (Life Technologies, InvitrogenTM, catalog number: 15240062)
- Recombinant hTGFβ1 (R&D Systems, catalog number: 100-B ) (see Recipes)
- Recombinant mIL-2 (R&D Systems, catalog number: 402-ML ) (see Recipes)
- NA/LE Hamster anti-mouse CD3ε monoclonal antibody (clone 145-2C11) (BD, catalog number: 553057 )
Equipment
- One milliliter syringe (BD, catalog number: 309628 )
- Sterile scissors
- P60 petri dishes (Greiner Bio-One GmbH, catalog number: 628160 )
- Sterile 70-μm filter (BD, catalog number: 352350 )
- Syringe filter (0.22 μm) (Corning, catalog number: 431219 )
- FACSAria cell sorter
- FACSAria II (BD, catalog number: 642510 )
- Airstream Class II BSC (ESCO Corporation)
- 15 ml conical tubes (BD, catalog number: 352095 )
- 5 ml polystyrene round bottom tubes (BD, catalog number: 352052 )
- Twenty four well plates (Greiner Bio-One GmbH, catalog number: 662160 )
- Centrifuge 5810R (Eppendorf, catalog number: 5811 000,622 )
- Dynal MCP-L (Life Technologies, InvitrogenTM, catalog number: 120.21D )
- CO2 incubator
Procedure
- Isolation of CD4+CD62L+ T naïve cells
- Prepare a single cell suspension from mouse spleens. Disrupt the spleen with the plunger of a 1 ml syringe against a 70-μm filter in a petri dish filled with 2 ml of RPMI.
- Centrifuge single cell suspensions in 15-ml conical tubes for 8 min at no more than 300 x g.
- Re-suspend the splenocytes with 5 ml of ACK buffer and incubate 5 min at RT. Dilute it with PBS and centrifuge for 8 min at no more than 300 x g. Re-suspend cell pellet in FACS buffer and count cell number. Normally, each spleen yields between 80-100 x 106 splenocytes.
- Purification of CD4+ T cells by negative selection using Dynal® Mouse CD4 Cell Negative Isolation Kit is thoroughly detailed in the protocol provided by manufacturer. Protocol yield is usually 20-25% of spleen cells (http://tools.invitrogen.com/content/sfs/manuals/dynabeads_untouched_ms_CD4_man.pdf).
- After CD4+ T cells isolation adjust the cell concentration by centrifugation (8 min at 300 x g) and dilution in FACS buffer to 4 x 107/ml and proceed to CD4 and CD62L staining.
- Use 0.2 μg of APC-conjugated anti-CD4 antibody and 0.3 μg of PE-conjugated anti-CD62L antibody per 200 μl of CD4+ T cells suspension. Incubate 30 min at 4 °C in the dark.
- Wash cells with FACS buffer, centrifuge for 8 min at no more than 300 x g and re-suspend cell pellet with FACS buffer at a concentration of 3 x 107/ml.
- Using a FACSAria cell sorter proceed to the selection and sorting of the CD4+CD62Lhigh population. Exclude cell doublets using FSC-H vs. FSC-W and SSC-H vs. SSC-W dot plots. The total percentage should be between 60-70% for Balb/c mice strain and 50-60% for C57Bl/6 mice strain. Flow rate is recommended to be adjusted around 1-3. Sort precision could be set to “yield”. One should expect around 10 x 106 and 7 x 106 of CD4 T naïve cells per Balb/c and C57Bl/6 spleen, respectively.
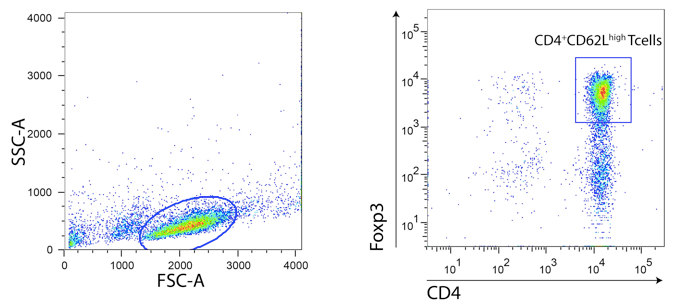
- Use 15 ml conical tubes to collect sorted population with 2.5 ml of collection medium. Prior to use, vortex tubes so that tube walls will be covered by a thin layer of fluid thus avoiding cell death when cells are deflected to the tube.
- Keep the sorted population on ice.
- Prepare a single cell suspension from mouse spleens. Disrupt the spleen with the plunger of a 1 ml syringe against a 70-μm filter in a petri dish filled with 2 ml of RPMI.
- Conditioned media from tumor cells
- This step can be performed at any time prior to the Treg differentiation protocol.
- Plate the chosen tumor cells in P60 dishes at 50% confluence with 2 ml of serum free media. Incubate for 18 h at 37 °C with 5% CO2 and then collect conditioned media. Filter with 0.22 μm syringe filter, aliquot into 200 µl samples and store at -70 °C.
- This step can be performed at any time prior to the Treg differentiation protocol.
- Anti-CD3 coating of 24-well plates
- Prepare a 5 μg/ml solution of anti-CD3ε from the stock of CD3ε antibody (1 mg/ml) in sterile PBS and vortex. For 24-well plates use 150 μl per well.
- Incubate at 37 °C in a humidified atmosphere for at least 2 h.
- Before use rinse wells with PBS and aspirate twice.
- Prepare a 5 μg/ml solution of anti-CD3ε from the stock of CD3ε antibody (1 mg/ml) in sterile PBS and vortex. For 24-well plates use 150 μl per well.
- Conversion of naïve T cells to Treg in the presence of tumor cells conditioned media
- The stimuli indispensable for Treg conversion are TGFβ1 and IL-2. To asses Gal-1 fine-tuning of Treg conversion frequency it is necessary to use a limiting concentration of the former. TGFβ1 limitation has shown to be more efficient at modulating Treg differentiation. Adjust naïve T cells concentration to 1 x 106/ml in serum free-RPMI supplemented with 1-2 ng/ml hTGFβ1, 100 U/ml mIL-2, 1 μg/ml CD28 mAb and antibiotic-antimycotic.
- Plate 1 ml of naïve T cell suspension per well in anti CD3-coated 24-well plates.
- Add conditioned media (CM) tumor cells. It is suggested to determine dose-dependent responses to the CM. Dilutions ranging from 1:10 to 1:100 are recommended.
- Incubate at 37 °C with 5% CO2 for 4 days. More days will only result in an increased cell death.
- Asses Treg frequency by flow cytometry after staining of CD4, CD25 and Foxp3.
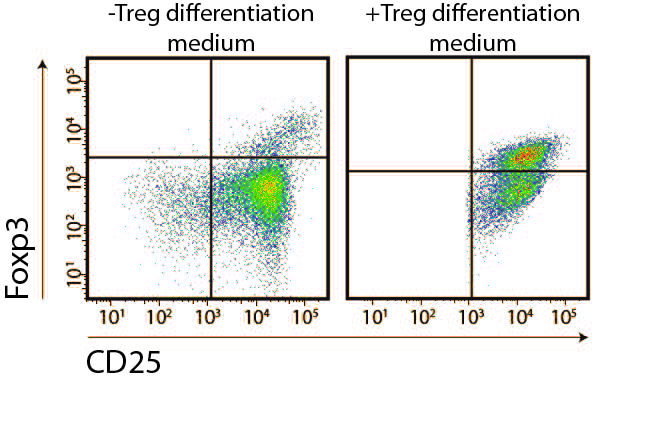
- The stimuli indispensable for Treg conversion are TGFβ1 and IL-2. To asses Gal-1 fine-tuning of Treg conversion frequency it is necessary to use a limiting concentration of the former. TGFβ1 limitation has shown to be more efficient at modulating Treg differentiation. Adjust naïve T cells concentration to 1 x 106/ml in serum free-RPMI supplemented with 1-2 ng/ml hTGFβ1, 100 U/ml mIL-2, 1 μg/ml CD28 mAb and antibiotic-antimycotic.
- Treg staining
- Staining of CD4 and CD25 molecules are performed for 30 min in the dark at 4 °C. Per 2 x 106 cells use 0.03 μg of APC-conjugated CD4 antibody and 0.075 μg of AlexaFluor 488-conjugated CD25 in 100 μl of FACS buffer.
- Wash cells and centrifuge for 8 min at no more than 300 x g. Fix and permeabilize cells using Fix/Perm buffer in 100 μl for 30 min to 18 h in the dark at 4 °C.
- Wash cells with 1x Permeabilization Buffer. Foxp3 staining is performed in 100 μl 1x Permeabilization Buffer using 0.225 μg PE-conjugated Foxp3 antibody for 1 h at 4 °C in the dark.
- Wash cells with 1x Permeabilization Buffer, centrifuge for 10 min at 300 x g and re-suspend in FACS buffer.
- For flow cytometry analysis a two-laser cytometer must be used and 5 additional tubes containing the appropriate compensation samples should be considered. It is highly recommended to exclude cell doublets using FSC-H vs. FSC-W and SSC-H vs. SSC-W dot plots.
- Staining of CD4 and CD25 molecules are performed for 30 min in the dark at 4 °C. Per 2 x 106 cells use 0.03 μg of APC-conjugated CD4 antibody and 0.075 μg of AlexaFluor 488-conjugated CD25 in 100 μl of FACS buffer.
Recipes
- Phosphate buffer saline (PBS)
136 mM NaCl
8.2 mM Na2HPO4
1.5 mM KH2PO4
2.7 mM KCl (pH 7.4)
- Sterile red blood lysis buffer (ACK buffer)
150 mM NH4Cl
10 mM KHCO3
0.1 mM EDTA
Resuspend in distilled H2O
Filter sterilize (0.45 μm)
Stored at 4 °C
- FACS buffer
PBS with 0.1% BSA and 2 mM EDTA
- Sorted cells collection medium
RPMI 1640 supplemented with 20% FBS
- Recombinant hTGFβ1
Dissolved in phosphate buffer saline (PBS) (pH 7.4) to a working dilution 30 µg/ml Stored in aliquots at -70 °C
- Recombinant mIL-2
Dissolved in PBS to a working dilution of 10 μg/ml
Stored in aliquots at -70 °C
Note: Avoid repeated freeze-thaw cycles as it may lead to loss of activity.
Acknowledgments
This protocol is based in the original work published in Dalotto-Moreno et al. (2013). This work was supported by grants from Agencia Nacional de Promoción Científica y Técnica Argentina (ANPCyT; PICT 2007-093 to M.S. and 2010-870 to G.A.R. and Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET; PIP 2010-2012 to M.S. and G.A.R.), Fundación Sales to G.A.R. The authors wish to express special thanks to María Rosa Morales for animal technical help.
References
- Curiel, T. J., Coukos, G., Zou, L., Alvarez, X., Cheng, P., Mottram, P., Evdemon-Hogan, M., Conejo-Garcia, J. R., Zhang, L., Burow, M., Zhu, Y., Wei, S., Kryczek, I., Daniel, B., Gordon, A., Myers, L., Lackner, A., Disis, M. L., Knutson, K. L., Chen, L. and Zou, W. (2004). Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 10(9): 942-949.
- Dalotto-Moreno, T., Croci, D. O., Cerliani, J. P., Martinez-Allo, V. C., Dergan-Dylon, S., Mendez-Huergo, S. P., Stupirski, J. C., Mazal, D., Osinaga, E., Toscano, M. A., Sundblad, V., Rabinovich, G. A. and Salatino, M. (2013). Targeting galectin-1 overcomes breast cancer-associated immunosuppression and prevents metastatic disease. Cancer Res 73(3): 1107-1117.
- Sakaguchi, S. (2004). Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol 22: 531-562.
- Zou, W. (2006). Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol 6(4): 295-307.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Dalotto-Moreno, T., Rabinovich, G. A. and Salatino, M. (2014). In vitro Regulatory T cells Differentiation From Naïve T Cells. Bio-protocol 4(6): e1075. DOI: 10.21769/BioProtoc.1075.
Category
Cancer Biology > Tumor immunology > Cell biology assays > Cell isolation and culture
Cell Biology > Cell viability > Cell proliferation
Immunology > Immune cell isolation > Lymphocyte
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











