- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measuring Genetic Robustness in Vesicular Stomatitis Virus
Published: Vol 4, Iss 6, Mar 20, 2014 DOI: 10.21769/BioProtoc.1073 Views: 9332
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Detachment Procedure of Bacteria from Atmospheric Particles for Flow-cytometry Counting
Carolina M. Araya [...] Isabel Reche
Jun 20, 2019 5592 Views

Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 3038 Views
Abstract
Genetic robustness is the ability of a genome to incorporate mutations with the result of no fitness changes. Thus, more robust viruses have an increased neutral mutation rate. This property is particularly important in RNA viruses due to their high mutation rates. The most direct way of measuring robustness in vesicular stomatitis virus (VSV) is to carry out clonal analysis of populations: randomly isolating individual VSV strains (plaques), measuring the fitness of each one and generating fitness distributions (Novella et al., 2010). A second possibility is to carry out multiple replicates of repeated plaque-to-plaque passages, determining fitness in progeny populations and generating fitness distributions (Novella et al., 2010). Depending on the expected differences, the former may require hundreds of determinations, while the latter may require tens of determinations. A third approach consists of increasing the mutation rate of populations under analysis to magnify any differences that may exist and, instead of measuring fitness, measuring survival (Novella et al., 2013). One caveat of this method is that changes in survival can also be explained by changes in polymerase fidelity. For that reason, it is important to perform complementary experiments, in this case quantifying mutant frequency.
Materials and Reagents
- Test and reference VSV strains
Note : The former is the strain under investigation, the latter is the control (typically the progenitor).
- Baby hamster kidney cells (BHK-21)
- 10x Trypsin/EDTA (Life Technologies, Gibco®, catalog number: 15400 )
- I1 Monoclonal antibody (I1Mab) hybridoma (Holland et al., 1991) (ATCC, catalog number: CRL-2700 )
Note: This antibody recognizes the G glycoprotein of VSV.
- I14 Mab (Holland et al., 1991) (https://www.kerafast.com/p-172-hybridoma-ie9f9-i14.aspx)
Note: This antibody recognizes the G glycoprotein of VSV.
- Proteose peptone No. 3 (PP3) (BD DifcoTM, catalog number: 212230 ) (12 g/L in dH2O, autoclaved)
- Bovine Calf Serum (BCS) (Life Technologies, Gibco®, catalog number: 16170-078 )
Note: BCS is a good choice to carry out plaque assays and it is a lot cheaper than FBS.
- Fetal Bovine Serum (FBS) (Life Technologies, Gibco®, catalog number: 10437 )
- 10% CO2
- 5-Fluorouracyl (5-FU) (Sigma-Aldrich, catalog number: F-6627 ) (10 mg/ml in ethanol, filtered)
- Agarose (Lonza, SeqplaqueTM GTGTM, catalog number: 50111 ) (40 g/L in dH2O, autoclaved)
- 4.2% bicarbonate
- Penicillin/streptomycin mixture (Mediatech, Cellgro®, catalog number: 30-004-Cl )
- Saline solution (see Recipes)
- Minimal Essential Medium with Hank’s salts (MEM-H) (Mediatech, Cellgro®, catalog number: 50-019-PB ) (see Recipes)
- Crystal violet solution (see Recipes)
Equipment
- T25 plug-seal flasks (CytoOne®, catalog number: CC7682-4325)
- 5 ml and 25 ml pipetes
- Plugged Pasteur pipetes
- 1.5 ml tubes
- Type II biosafety hood
- 37 °C, CO2 cell culture incubator
- Transilluminator (optional)
Procedure
- Make I1 Mab stock and titrate I1 and I14 antibodies; use at enough concentration to produce full inhibition of wild type. Alternatively, I1 Mab can be purchased from Kerafast (https://www.kerafast.com/p-171-hybridoma-8g5f11-i1.aspx).
Notes:- Because of high mutation rates leading to high frequency of antibody-resistance mutants, viral stocks can never be fully neutralized. The goal of Mab titration is to produce plaque assays with less than 10-4 (if viral stocks are neutralized) or less than 10-3 (if the Mab is added to the overlay medium) mutants/wt.
- Antibodies other than I14 (https://www.kerafast.com) can be used to calculate mutant frequencies.
- Because of high mutation rates leading to high frequency of antibody-resistance mutants, viral stocks can never be fully neutralized. The goal of Mab titration is to produce plaque assays with less than 10-4 (if viral stocks are neutralized) or less than 10-3 (if the Mab is added to the overlay medium) mutants/wt.
- BHK-21 cells are washed twice with saline solution, trypsinized and 0.8-1.0 x 105 are seeded in T25 flasks with MEM-H supplemented with 7% BCS and 0.06% PP3. The flasks are gassed for 2 sec with 10% CO2 using a plugged Pasteur pipette. Caps are locked.
- The cells are incubated for 24 h to produce monolayers 90% confluent.
- On day 2 cell monolayers are treated with no mutagen (mock) or with the mutagen 5-FU at a range of concentrations between 1 and 100 μg/ml for 6 h at 37 °C (5, 10, 25, 50 and 100 μg/ml).
- Dilute viral stocks in MEM-H+FBS as needed to produce a 106 plaque forming units (PFU)/ml solution.
- Use 200 μl to infect a BHK-21 monolayer for each 5-FU concentration. Incubate 10 min at room temperature (RT) and 40 min at 37 °C. Add 5 ml of MEM-H+FBS+5-FU (Figure 1, right).
- From the 106 PFU/ml solution carry out additional, 10-fold, 100-fold and 1000-fold dilutions. Use each of the three dilutions (105, 104 and 103 PFU/ml solutions) to infect at least 3 BHK-21 monolayers with 200 μl of the dilution to carry out plaque assay in the presence of I1Mab. Do not neutralize the mixture with I14Mab prior to infection, as it will result in incorrect data due to phenotypic mixing and hiding (Valcarcel and Ortin, 1989; Holland et al., 1989). Incubate 10 min at room temperature (RT) and 40 min at 37 °C. Prewarm a mixture of MEM-H+FBS and I14Mab, add agarose to a final concentration of 0.2%, and add 5 ml of mixture to flasks (Figure 1, left).
Note: Even if the virus stock has a known concentration it is recommended that a titration is done in parallel by triplicate plaque assay Figure 1, top left).
Figure 1. Flowchart showing the infections needed to calculate the frequency of I1 MARM and the sensitivity to 5-FU. Details are given in the text.
- Use 200 μl to infect a BHK-21 monolayer for each 5-FU concentration. Incubate 10 min at room temperature (RT) and 40 min at 37 °C. Add 5 ml of MEM-H+FBS+5-FU (Figure 1, right).
- Incubate mutagenized infections and plaque assays for 24 h at 37 °C (Figure 1).
- Develop all plaque assays (Figure 1). Because of the low agarose concentration there is no need to fix the cells. Just let the medium overlay slide out and add 2-3 ml of crystal violet solution. Wait for 5 min at RT, discard crystal violet and rinse with tap water.
- Count plaques and calculate mutant frequency by dividing Monoclonal antibody resistant mutant (MARM) titers (Figure 1B) by wt titers (Figure 1A).
- Check virus replication by examining monolayers under the microscope. When cytopatic effect is > 90%, recover mutagenized populations.
Notes:- For lower FU concentrations 24 h are usually sufficient, but for FU concentrations over 30 μg/ml it may take up to three days.
- Samples may be frozen so titrations can be carried out for the complete set of mutagenized samples in a single assay.
- For lower FU concentrations 24 h are usually sufficient, but for FU concentrations over 30 μg/ml it may take up to three days.
- Carry out titrations of control and mutagenized viral yields. To improve statistical significante it is recommended to do duplicate or triplicate plaque assays for each dilution. Allow 24-36 h of incubation.
- Develop plaque assays with crystal violet and count claque (Figure 1C).
- Robustness can be calculated as the slope of the regression of log transformed normalized viral titers vs. square-root transformed 5-FU concentration (Figure 2). The more robust the strain is the closer to a slope of 1.
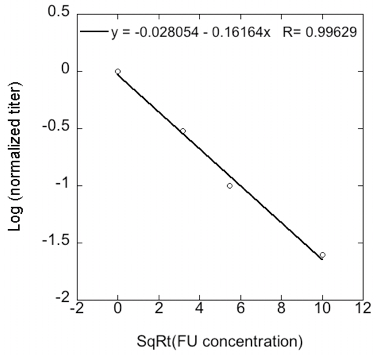
Figure 2. To calculate robustness the log-transformed viral titer is represented against the square root of mutagen concentration. Robustness is the slope of the regression (shown at the top of the graph).
- To test whether changes in survival are the result of changes in the overall mutation rate, and not in the neutral mutation rate, mutant frequencies must be compared. When all other environmental parameters are the same, mutant frequencies correlate with mutation rates. If mutant frequency and survival to mutagenesis correlate inversely, the latter may not represent robustness. In such case, a clonal analysis is indicated.
Recipes
- Saline solution
Add 7 g of NaCl to dH2O to 1,000 ml
Autoclave 20 min
Stored at RT
- MEM-H
Mix powder with 9.65 L of dH2O
Add:
250 ml of 4.2% bicarbonate
100 ml of penicillin/streptomycin mixture
Filter and aliquote in 1 L bottles. At this time the medium can be stored in the fridge.
When ready for use add:
70 ml of serum (FEB or BCS)
5 ml of 12% PP3 (if needed)
Notes:- Making medium is worth it if there is a large volume of cell culture performed in the laboratory. Ready-to-use MEM-E can also be purchased from several companies.
- MEM with Earls salts (MEM-E) is also a good choice, but requires a CO2 incubator.
- Making medium is worth it if there is a large volume of cell culture performed in the laboratory. Ready-to-use MEM-E can also be purchased from several companies.
- Crystal violet solution
Mix 750 ml H2O, 250 ml ethanol and 5 g crystal violet
Stir until dissolved and stored at RT
Acknowledgments
This protocol was originally published in Novella et al. (2013). Work was supported by funds from the University of Toledo to ISN.
References
- Holland, J. J., de la Torre, J. C., Clarke, D. K. and Duarte, E. (1991). Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J Virol 65(6): 2960-2967.
- Holland, J. J., de la Torre, J. C., Steinhauer, D. A., Clarke, D., Duarte, E. and Domingo, E. (1989). Virus mutation frequencies can be greatly underestimated by monoclonal antibody neutralization of virions. J Virol 63(12): 5030-5036.
- Novella, I. S., Presloid, J. B., Zhou, T., Smith-Tsurkan, S. D., Ebendick-Corpus, B. E., Dutta, R. N., Lust, K. L. and Wilke, C. O. (2010). Genomic evolution of vesicular stomatitis virus strains with differences in adaptability. J Virol 84(10): 4960-4968.
- Novella, I. S., Presloid, J. B., Beech, C. and Wilke, C. O. (2013). Congruent evolution of fitness and genetic robustness in vesicular stomatitis virus. J Virol 87(9): 4923-4928.
- Valcarcel, J. and Ortin, J. (1989). Phenotypic hiding: the carryover of mutations in RNA viruses as shown by detection of mar mutants in influenza virus. J Virol 63(9): 4107-4109.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Novella, I. S. (2014). Measuring Genetic Robustness in Vesicular Stomatitis Virus. Bio-protocol 4(6): e1073. DOI: 10.21769/BioProtoc.1073.
Category
Microbiology > Microbial genetics > Mutagenesis
Molecular Biology > RNA > RNA sequencing
Microbiology > Microbial cell biology > Cell isolation and culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link