- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Cellular Translational Reporter Assay
Published: Vol 4, Iss 6, Mar 20, 2014 DOI: 10.21769/BioProtoc.1068 Views: 11775
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In Vitro Analysis of Stalled Ribosomes using Puromycin Incorporation
MaKenzie R. Scarpitti and Michael G. Kearse
Aug 20, 2023 3338 Views

Immunoprecipitation of Reporter Nascent Chains from Active Ribosomes to Study Translation Efficiency
Roberta Cacioppo and Catherine Lindon
Sep 20, 2023 2127 Views

Metabolic RNA Labeling and Translating Ribosome Affinity Purification for Measurement of Nascent RNA Translation
Hirotatsu Imai and Akio Yamashita
Oct 20, 2024 2522 Views
Abstract
The method described here allows measuring the effect of exogenously introduced modifications to in vitro-transcribed mRNA on the translation in cells. Using cells derived from knockout mice and control littermates, this method enables to compare the results in the presence or absence of specific gene products. In our lab, we used this protocol to check whether the exogenous addition of 5’ capping and 2’-O methylation to in vitro-mRNA affects the translational efficiency. Here we describe the details of our experiments.
Materials and Reagents
- Mouse embryonic fibroblasts (prepared from day 14.5 embryos)
- Vector pGL4.14 (Luc2 encoding vector) (Promega Corporation, catalog number: E6691 )
- Primers (Invitrogen custom DNA primers)
- 5’-TAATACGACTCACTATAGGCCACCATGGAAGATGCCAAAAA-3’ (The T7 class III promoter sequence is underlined.)
- 5’-TACCACATTTGTAGAGGTTTTACTTGCTTT-3’
- 5’-TAATACGACTCACTATAGGCCACCATGGAAGATGCCAAAAA-3’ (The T7 class III promoter sequence is underlined.)
- rTaq DNA polymerase (TOYOBO, catalog number: TAP-211 )
- Agarose gel
- Ethidium bromide
- Illustra GFX PCR and Gel Band Purification Kit (GE, catalog number: 28-9034-71 )
- MEGAScript In vitro transcription Kit (Life Technologies, Ambion®, catalog number: AM1333 )
Note: Nuclease-free Water and LiCl Precipitation Solution are included in the kit.
- 80% ethanol
- ScriptCap m7G capping system (EpiCentre, catalog number: SCCE0610 )
- ScriptCap 2’-O-Methyltransferase Kit (EpiCentre, catalog number: SCMT0610 )
- RNeasy Mini Kit (QIAGEN, catalog number: 74104 )
- Opti-MEM I reduced serum medium (Life Technologies, Gibco®, catalog number: 31985-070 )
- D-PBS(-) (Nacalai Tesque, catalog number: 14249-95 )
- Lipofectamine 2000 DNA Transfection Reagent (Life Technologies, catalog number: 11668-019 )
- Dual-luciferase reporter assay system (Promega Corporation, catalog number: E1960 )
- BCA protein assay reagent (Thermo Fisher Scientific, catalog number: 23227 )
Equipment
- Cell scraper
- GeneAmp PCR system 9700 (Applied Biosystems®)
- Centrifugal Concentrator CC105 (TOMY)
- Lumat LB 9507 Luminometer (Berthold Technologies)
- Model 680 Microplate Reader (Bio-Rad Laboratories)
Procedure
- Subcloning of Luc2 cDNA
- Prepare the polymerase chain reaction mix.
Mix following components on ice:
0.2 μl of rTaq DNA polymerase
5 μl of 10x Buffer (+Mg) for rTaq
5 μl of dNTPs (2 mM each)
4 μl of primers (10 mM each of Forward and Reverse primers)
1 μl of vector pGL4.14 (100 ng)
35 μl of water
- Run the polymerase chain reaction.
Place the reaction mix to the GeneAmp PCR system 9700.
PCR program:
Step 1: 94 °C 5 min
Step 2: 94 °C 30 sec
Step 3: 60 °C 2 min
Step 4: 74 °C 1 min (repeat steps 2-4 for 35 times)
Step 5: 74 °C 10 min
- Purification of PCR amplified fragments.
- Electrophorese the PCR products on 1% of agarose gel.
- Stain Gel with Ethidium Bromide.
- Cut the gel region including an amplified DNA fragment (approximately 1.6-kbp).
- Purify PCR amplified DNA from gels using illustra GFX PCR and Gel Band Purification Kit.
Note: In this process, we elute DNA fragment with 30 μl of Nuclease-free Water (8 μl aliquots of the eluent are used as a template DNA for following in vitro transcription).
- Electrophorese the PCR products on 1% of agarose gel.
- Prepare the polymerase chain reaction mix.
- In vitro transcription of Luciferase mRNA under the control of T7 promoter
- Prepare the in vitro transcription mix.
Mix following components at room temperature:
8 μl of Template DNA
2 μl of 10x Reaction Buffer
2 μl of 10 mM ATP
2 μl of 10 mM CTP
2 μl of 10 mM UTP
2 μl of 10 mM GTP
2 μl of T7 Enzyme Mix
Mix thoroughly with pipetting.
- Incubate at 37 °C, 4 h.
- Add 2 μl of TURBO DNase (a component of MEGAScript) to digest the rest template DNA and incubate 15 min at 37 °C on the block incubator.
- RNA extraction with lithium chloride (LiCl) precipitation.
- Add 30 μl of nuclease-free water and 30 μl of LiCl precipitation solution to the in vitro transcription products.
- Mix thoroughly with vortex mixer. Chill for 1 h at -20 °C.
- Centrifuge at 4 °C for 15 min at 15,000 rpm.
- Discard the supernatants, and wash the pellet with 1 ml of 80% ethanol.
- Centrifuge at 4 °C for 15 min at 15,000 rpm.
- Discard the supernatants, and dry the pellet with centrifugal concentrator CC105 for 5 min.
- Resuspend the RNA in 20 μl of Nuclease-free Water, and stored at -80 °C.
- Add 30 μl of nuclease-free water and 30 μl of LiCl precipitation solution to the in vitro transcription products.
- Prepare the in vitro transcription mix.
- Enzymatic modification of RNA 5’ end
- Add m7G cap, or m7G cap and 2’-O methylation to the in vitro-transcribed RNA with ScriptCap system.
- Adjust the volume of transcribed RNA (50 μg) to 67 μl with Nuclease-free Water.
- Denature the RNA at 65 °C for 10 min, then transfer the tube immediately to ice.
- While the RNA is denaturing, mix following components.
10 μl of 10x ScriptCap Capping Buffer
10 μl of 10 mM GTP
2.5 μl of 20 mM SAM
2.5 μl of ScriptGuard RNase Inhibitor
4 μl of ScriptCap 2’-O-Methyltrasferase (100 U/μl) for 2’-O methylated 5’ capped RNA or Nuclease-free Water for 2’-O unmethylated 5’ capped RNA
- Add 4 μl of ScriptCap Capping Enzyme (10 U/μl) and 67 μl of denatured RNA to the reaction cocktail from step C1c.
- Incubate at 37 °C for 2 h.
- Reaction products were purified with an RNeasy Mini Kit.
- Adjust the volume of transcribed RNA (50 μg) to 67 μl with Nuclease-free Water.
- Add m7G cap, or m7G cap and 2’-O methylation to the in vitro-transcribed RNA with ScriptCap system.
- RNA transfection with mouse embryonic fibroblasts
- Seed a 35 mm diameter dish with 2 x 105 mouse embryonic fibroblasts in 1 ml DMEM, and incubate 24 h.
- Transfect RNA using Lipofectamine 2000.
- Dilute 10 μl of Lipofectamine 2000 DNA Transfection Reagent in 240 μl of Opti-MEM medium per sample.
- Dilute 2 μg of luciferase RNA in 250 μl of Opti-MEM medium.
- Add 250 μl of diluted Lipofectamine 2000 DNA Transfection Reagent to 250 μl of diluted Luciferase RNA. Mix thoroughly.
- Incubate at room temperature for 15 min.
- Add RNA-lipid complex to the mouse embryonic fibroblasts, and incubate for 6 h.
- Dilute 10 μl of Lipofectamine 2000 DNA Transfection Reagent in 240 μl of Opti-MEM medium per sample.
- Seed a 35 mm diameter dish with 2 x 105 mouse embryonic fibroblasts in 1 ml DMEM, and incubate 24 h.
- Luciferase translational reporter assays
- Preparation of cell lysates.
- After incubation for 6 h, wash the RNA-transfected mouse embryonic fibroblasts with D-PBS(-) twice.
- Add 100 μl of 1x passive lysis buffer and harvest cells with cell scraper.
- Centrifuge at 4 °C for 15 min at 15,000 rpm.
- Determine the protein concentration in 5 μl aliquots of the supernatants by BCA Protein Assay.
- Add equal protein amount (determined by BCA protein assay) of aliquots of the supernatants to 50 μl of Luciferase Assay Reagent II (LAR II).
- Immediately, measure the relative luciferase units (RLUs) on Lumat LB 9507 Luminometer according to the manufacturer’s instruction.
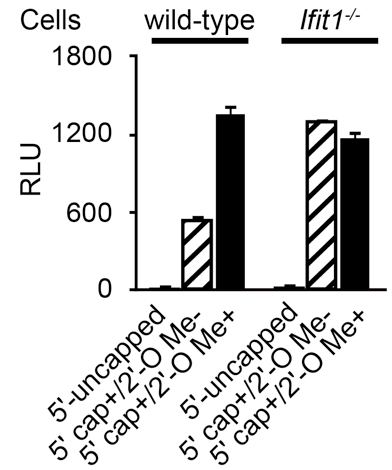
Figure 1. Luciferase activity of introduced RNAs. Wild-type and Ifit1-deficient (Ifit1-/-) mouse embryonic fibroblasts were transiently transfected with three different types of luciferase mRNAs (5’-uncapped, 5’ capped but 2’-O unmethylated; 5’ cap+/2’-O Me-, 5’ capped and 2’-O methylated; 5’ cap+/2’-O Me+). Luciferase activities are shown as relative light units (RLU), and the numbers of RLU were normalized by the concentrations of proteins determined in step E1d. Data are shown as means ± SDs of triplicate samples. These data show that Ifit1 selectively inhibits the translation of 5’ capped but 2’-O unmethylated (5’ cap+/2’-O Me-) luciferase mRNA.
- After incubation for 6 h, wash the RNA-transfected mouse embryonic fibroblasts with D-PBS(-) twice.
- Preparation of cell lysates.
Acknowledgments
This work was adapted from the following paper: Kimura et al. (2013). This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology, the Japan Science and Technology Agency, and by the Ministry of Health, Labour and Welfare.
References
- Kimura, T., Katoh, H., Kayama, H., Saiga, H., Okuyama, M., Okamoto, T., Umemoto, E., Matsuura, Y., Yamamoto, M. and Takeda, K. (2013). Ifit1 inhibits Japanese encephalitis virus replication through binding to 5′ capped 2′-O unmethylated RNA. J Virol 87(18): 9997-10003.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kimura, T. and Takeda, K. (2014). Cellular Translational Reporter Assay. Bio-protocol 4(6): e1068. DOI: 10.21769/BioProtoc.1068.
Category
Biochemistry > RNA > mRNA translation
Microbiology > Microbial biochemistry
Molecular Biology > RNA > mRNA translation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








