- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Lysosomal Amino Acid Efflux Assay
Published: Vol 4, Iss 4, Feb 20, 2014 DOI: 10.21769/BioProtoc.1048 Views: 10442
Reviewed by: Lin FangFanglian HeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Polyamine Transport Assay Using Reconstituted Yeast Membranes
Sarah van Veen [...] Peter Vangheluwe
Jan 20, 2021 4504 Views

An in vitro Blood-brain Barrier Model to Study the Penetration of Nanoparticles
Sezin Aday [...] Nitin Joshi
Feb 20, 2022 11792 Views

A Robust Nanoparticle-based Magnetic Separation Method for Intact Lysosomes
The Son Le [...] Shinya Maenosono
Jul 5, 2022 3284 Views
Abstract
As the cellular “recycling” organelle, lysosomes break down proteins into amino acids, which are then transported into cytosol for reuse by various amino acid transporters. The transport rate of an amino acid is presumably regulated by cellular conditions such as organelle pH, membrane potential and metabolic states. Because of their intracellular localization and the relative inaccessibility, lysosomal amino acid transporters have been studied largely via indirect measurements. Using lysosome purification and 14C-labeled amino acids, this protocol provides a method to measure the efficiency of specific amino acid transporters on lysosomes.
Materials and Reagents
- Mouse liver
- 14C-labeled amino acids (PerkinElmer) (select the amino acid of your interest)
- 3 N methanolic HCl (Sigma-Aldrich, catalog number: 33051 )
- Anhydrous methanol (Sigma-Aldrich, catalog number: 322415 )
- Sucrose
- HEPES
- MOPS
- Na2EDTA
- KCl
- MgCl2
- ATP-Mg
- Phosphate buffered saline (PBS)
- Scintillation cocktail (Research Products International, catalog number: 111175 )
- Protein assay kit (Bio-Rad Laboratories, catalog number: 500-0112 )
- Homogenization buffer (HB) (see Recipes)
- High sucrose buffer (see Recipes)
- Uptake buffer (see Recipes)
- Efflux buffer (see Recipes)
Equipment
- Nitrogen gas tank (with regulator and connecting tube)
- 25-ml glass conical flask
- 1.5 ml and 15 ml tubes
- 30 G needles
- Dounce homogenizer
- High speed centrifuge (up to 25,000 x g) with temperature control
- Glassfiber filters (Grade GF/F) (Whatman, catalog number: 1825-025 )
- Vacuum filtration manifold (EMD Millipore, model: 1225 )
- Liquid scintillation counter (Beckman Coulter, model: LS6500 )
- Personal protective equipment for working with radioactive materials
Procedure
Note: This protocol involves 14C-labeled amino acids. The uses of radioactive isotopes and the necessary safety precautions to be taken should follow institutional regulations.
- Preparation of amino acid methyl esters
- Transfer 12.5 µCi 14C-labeled amino acid into a 25-ml glass conical flask and let it dry under stream of nitrogen.
Note: The drying time depends on the speed of nitrogen flow and is about 10 min.
- Add 3 ml 3 N methanolic HCl into the flask and incubate for 24 h at room temperature. This will transform amino acid into amino acid methyl ester.
Note: Seal the opening of the flask with parafilm to prevent evaporation of methanolic HCl. The transformation efficiency can be measured with chromatography and high voltage electrophoresis (Reeves, 1979; Steinherz et al., 1982).
- Dry the resulted amino acid methyl ester under stream of nitrogen. Add 1 ml anhydrous methanol to dissolve the pellet and dry it again under stream of nitrogen.
Note: The drying time depends on the speed of nitrogen flow.
- Add 500 μl anhydrous methanol to dissolve the methyl ester. Bring down the volume to 250 μl through evaporation under stream of nitrogen.
- Shortly before efflux assay, transfer appropriate amount into a 1.5 ml tube. Dry under stream of nitrogen. Dissolve in uptake buffer (final concentration 0.005 μCi/μl).
Note: Amino acid methyl ester is not stable in aqueous solution.
- Transfer 12.5 µCi 14C-labeled amino acid into a 25-ml glass conical flask and let it dry under stream of nitrogen.
- Crude lysosome preparation
- To prepare crude lysosome preparation from mouse liver, follow animal use protocols and dissect out the liver after the animal is euthanized. Rinse in ice-cold HB.
Note: Liver from one adult mouse is enough for one assay. If you need to test multiple amino acids, calculate the total protein concentration you can get from one mouse liver and scale up the animal number.
- Transfer the liver into a Dounce homogenizer, add 8 ml ice-cold HB.
- Homogenize the liver on ice to break liver cells (20 strokes).
- Centrifuge the homogenate at 1,000 x g for 10 min at 4 °C; transfer the supernatant into a new centrifuge tube and keep on ice.
- Homogenize the pellet again in 7 ml HB (15 strokes); centrifuge at 1,000 x g for 10 min at 4 °C.
- Combine the supernatant from steps B4-5; centrifuge at 20,000 x g for 20 min at 4 °C.
- Resuspend the pellet in 10 ml 250 mM KCl (pH 7.4, buffered by 1 mM MOPS) and centrifuge at 1,600 x g for 10 min at 4 °C. Transfer the supernatant to a fresh tube. This step pellets most of the mitochondria.
- Dilute the supernatant by 2.5x volume high sucrose buffer and centrifuge at 25,000 x g for 10 min to spin down the crude lysosomes.
- Resuspend the crude lysosome pellet in 200 μl uptake buffer. Centrifuge at 600 x g for 2 min.
- Transfer the supernatant to a new tube and keep it on ice. Resuspend the pellet in 100 μl uptake buffer by passing through a 30 G needle for 30 times. Combine it with the supernatant. This will be the crude lysosome sample.
- Use a small aliquot of crude lysosome to measure total protein concentration with protein assay. Adjust the concentration to 2 mg protein-equivalent amount of lysosomes/ml. Keep the rest of the preparation on ice and do the efflux assay within 12 h. Longer storage is not recommended.
Note: If needed, lysosomes can be further purified from the above preparation using, for example, OptiPrep density gradient centrifugation (see References 4 and 5).
- To prepare crude lysosome preparation from mouse liver, follow animal use protocols and dissect out the liver after the animal is euthanized. Rinse in ice-cold HB.
- Amino acid efflux assay
- For each reaction, dispense 50 μl aliquot (containing 100 μg of protein-equivalent amount of lysosomes) into a 1.5 ml tube.
- To initiate amino acid ester loading, add 50 μl of amino acid methyl ester (0.25 μCi, in uptake buffer) into each lysosome aliquot tube. Gently mix by pipetting. Incubate at 37 °C for 30 min.
Note: The optimum loading time and temperature need to be experimentally determined for the amino acid and the type of lysososomes used.
- During the loading time, assemble the Whatman filters (GF/F) to vacuum filtration manifold. Rinse the filters with 5 ml PBS. Adjust the filtering speed to ~10 ml/min.
- At the end of loading, dilute the loading reaction 50x by transferring the 100 μl loading reaction into a 15 ml tube containing 5 ml pre-warmed (37 °C) efflux buffer; mix well and incubate at 37 °C. This will start the efflux.
- At the end of each efflux time point, combine a tube of efflux reaction (containing 5.1 ml) with 5 ml ice-cold PBS. Filter immediately through Whatman filter under vacuum; wash twice with 5 ml ice-cold PBS.
- For background count, incubate lysosomes and amino acid methyl ester separately (without loading). Mix and filter immediately after incubation.
- Soak the filter in scintillation cocktail overnight. Count using a scintillation counter.
- Subtract background from the isotope counts. Normalize the background-subtracted counts to that obtained at efflux time point 0. The normalized count at each time point indicates the portion of amino acid remaining in the lysosome and reflects the activity of the amino acid transporter. (See Figure 1 for an example.)
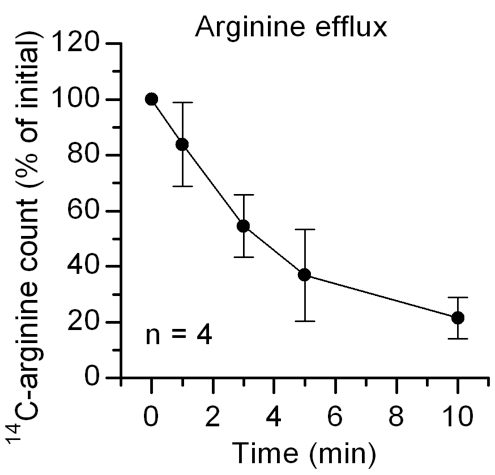
Figure 1. A representative amino acid efflux result using mouse crude lysosomes and 14C-labeled arginine. Data are presented as mean ± SEM.
- For each reaction, dispense 50 μl aliquot (containing 100 μg of protein-equivalent amount of lysosomes) into a 1.5 ml tube.
Recipes
- Homogenization buffer (HB)
250 mM sucrose
1 mM Na2EDTA
10 mM HEPES (pH 7.0)
- High sucrose buffer
450 mM sucrose
0.5 mM Na2EDTA
5 mM HEPES (pH 7.2)
- Uptake buffer
250 mM sucrose
5 mM MgCl2
20 mM HEPES (pH 7.0)
- Efflux buffer
250 mM sucrose
5 mM MgCl2
20 mM HEPES
2 mM ATP-Mg (pH 7.0)
Acknowledgments
This protocol was adapted from previously published papers (Cang et al., 2013; Reeves, 1979; Steinherz et al., 1982; Graham et al., 1982) and the technical bulletin of the lysosome isolation kit from Sigma-Alderich (Reference 5). We thank Dr. Yandong Zhou and Dr. Youngjun Seo for discussion and conducting the experiments, and Dr. Denia Ramirez-Montealegre for advice on the protocol. The development of this protocol was supported, in part, by funding from American Heart Association, NIH and the University of Pennsylvania Research Foundation.
References
- Cang, C., Zhou, Y., Navarro, B., Seo, Y. J., Aranda, K., Shi, L., Battaglia-Hsu, S., Nissim, I., Clapham, D. E. and Ren, D. (2013). mTOR regulates lysosomal ATP-sensitive two-pore Na(+) channels to adapt to metabolic state. Cell 152(4): 778-790.
- Graham, J., Ford, T. and Rickwood, D. (1994). The preparation of subcellular organelles from mouse liver in self-generated gradients of iodixanol. Anal Biochem 220(2): 367-373.
- Reeves, J. P. (1979). Accumulation of amino acids by lysosomes incubated with amino acid methyl esters. J Biol Chem 254(18): 8914-8921.
- Steinherz, R., Tietze, F., Raiford, D., Gahl, W. A. and Schulman, J. D. (1982). Patterns of amino acid efflux from isolated normal and cystinotic human leucocyte lysosomes. J Biol Chem 257(11): 6041-6049.
- Technical Bulletin of Lysosome Isolation Kit (LYSISO1). (Sigma-Aldrich)
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Cang, C. and Ren, D. (2014). Lysosomal Amino Acid Efflux Assay. Bio-protocol 4(4): e1048. DOI: 10.21769/BioProtoc.1048.
Category
Cell Biology > Organelle isolation > Lysosome
Cell Biology > Cell-based analysis > Transport
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










