Materials and Reagents
- Mouse spleen cells or ES-derived hematopoietic cells
- DNA extraction Kit: PerfectPure DNA Cultured Cell Kit (5 PRIME, catalog number: 2302000 )
- 10x PCR Buffer with KCl (Life Technologies, Applied Biosystems®, catalog number: 4338856 )
- MgCl2 (Life Technologies, Applied Biosystems®, catalog number: 4338856)
- Taq DNA polymerase (Life Technologies, Applied Biosystems®, catalog number: 4338856)
- dNTPs (Life Technologies, Applied Biosystems®, catalog number: 4338856)
- Primers (FASMAC)
The sequence of primers are as follows.
a. DHL(5’): GGAATTCG(AorC)TTTTTGT(CorG)AAGGGATCTACTACTGTG
b. Mu0(5’): CCGCATGCCAAGGCTAGCCTGAAAGATTACC
c. J3(3’): GTCTAGATTCTCACAAGAGTCCGATAGACCCTGG
- Agarose (UltraPureTM Agarose) (Life Technologies, InvitrogenTM, catalog number: 16500-100 )
- Ethidium bromide (Wako Pure Chemical Industries, catalog number: 315-90051 )
Equipment
- PCR Thermal Cycler (Veriti Thermal Cycler) (Life Technologies, Applied Biosystems®)
- Centrifuges (TOMY SEIKO, model: MX-150 )
- Electrophoresis apparatus (ADVANCE, Mupid-exU)
Procedure
- Genomic DNA was prepared for PCR by lysing mouse spleen cells (1-4 x 104) or ES-derived hematopoietic cells (3-5 x 105) in 75 µl elution solution. See the manufacturer's protocol (http://www.5prime.com/media/3415/perfectpure dna cultured cell manual_5prime_1064553_122010.pdf).
- 20 µl PCR reactions contained 5.5 µl template (82.5 ng or less), 10 mM Tris-HCl, 50 mM KCl, 2.0 mM MgCl2, 1 µM primers (25 mers), 200 µM dNTPs and 1 U Taq DNA polymerase.
- PCR program
a.94 °C – 2 min
b.94 °C – 1 min
c.60 °C – 1 min
d.72 °C – 1.75 min
e.Repeat steps b-d, 35x
f.72 °C – 10 min
- Half of each PCR products were electrophoresed on 1% agarose gels, and their amounts were evaluated by staining with ethidium bromide.
- DH-JH recombination was detected as amplified fragments of 1,033 bp, 716 bp and 333 bp using a primer DHL(5’) and J3(3’). Germline alleles were detected as an amplified fragment of 1,259 bp using a primer Mu0(5’) and J3(3’).
Results
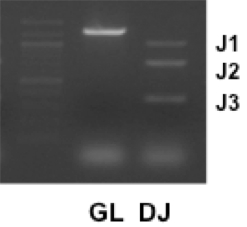
Figure 1.DNA PCR assays of germline (GL) or DH-JH rearranged Igh chain (DJ) genes were performed with mouse splenocytes. A PCR experiment using a primer DHL(5’) and J3(3’) can detect three types of DH-JH rearrangement (J1, J2, and J3) (Schlissel et al., 1991). All of three bands are present with successful DH-JH rearrangement. However, a J1 band is sometimes undetected in the ethidium bromide-based DNA-band visualization when the amount of template DNA is very small. The size marker was loaded in the left lane.
Acknowledgments
This protocol was adapted from a previously published paper by Schlissel et al. (1991). The representative data shown in the protocol was adapted from Satoh et al. (2013).
References
- Satoh, Y., Yokota, T., Sudo, T., Kondo, M., Lai, A., Kincade, P. W., Kouro, T., Iida, R., Kokame, K., Miyata, T., Habuchi, Y., Matsui, K., Tanaka, H., Matsumura, I., Oritani, K., Kohwi-Shigematsu, T. and Kanakura, Y. (2013). The Satb1 protein directs hematopoietic stem cell differentiation toward lymphoid lineages. Immunity 38(6): 1105-1115.
- Schlissel, M. S., Corcoran, L. M. and Baltimore, D. (1991). Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J Exp Med 173(3): 711-720.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
Category
Immunology > Antibody analysis > Antibody function
Molecular Biology > DNA > PCR
















