- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Surface Plasmon Resonance Method to Study HCV NS5B Inhibitors
Published: Vol 4, Iss 4, Feb 20, 2014 DOI: 10.21769/BioProtoc.1044 Views: 9493
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Kinetic Analysis of Monoclonal Antibody Binding to HIV-1 gp120-derived Hyperglycosylated Cores
Jidnyasa Ingale and Richard T Wyatt
Oct 5, 2015 8539 Views

Binding Affinity Quantifications of the Bacteriophage Mu DNA Modification Protein Mom Using Microscale Thermophoresis (MST)
Shubha Udupa [...] Shweta Karambelkar
Jul 20, 2022 3706 Views

Flotation Assay With Fluorescence Readout to Study Membrane Association of the Enteroviral Peripheral Membrane Protein 2C
Kasturika Shankar [...] Lars-Anders Carlson
Apr 5, 2025 1621 Views
Abstract
Surface Plasmon Resonance (SPR) technology is a well-established platform used to evaluate the kinetic parameters of protein-small molecule interactions. Below, we describe the use of the ProteOn XPR36 biosensor from Bio-Rad (Hercules, CA) to evaluate the binding of small molecule inhibitors to recombinant NS5B protein. The high pI (> 9) of this construct allows for chemical immobilization using HEPES-buffered saline at pH 7.5. This is in contrast to traditional biosensor protocols that use both low pH and ionic strength. The use of a more physiological buffer to immobilize this enzyme leads to improved surface activity.
Keywords: Surface Plasmon Resonance (SPR)Materials and Reagents
- HCV NS5B inhibitors
- Purified recombinant HCV NS5B ΔC21 soluble protein [cloned and purified according to Boyce et al. (2014) and Hung et al. (2011)]
- ProteOn GLH Sensor Chip (Bio-Rad Laboratories, catalog number: 176-5013 )
- HEPES solution (1 M) (Sigma-Aldrich, catalog number: H3537-1L )
- MgCl2 (1 M) (Sigma-Aldrich, catalog number: M1028-100ML )
- EDTA (0.5 M) (Sigma-Aldrich, catalog number: E7889-100ML )
- NaCl (5 M) (Sigma-Aldrich, catalog number: S6546-1L )
- KCl (2 M) (Life Technologies, Ambion®, catalog number: AM9640G )
- Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) (Sigma-Aldrich, catalog number: C4706-10G )
- Surfactant P20 (10% v/v) (General Electric Company, catalog number: BR-1000-54 )
- DMSO (Sigma-Aldrich, catalog number: 472301-100ML )
- ProteOn Amine Coupling Kit (Bio-Rad Laboratories, catalog number: 176-2410 ) containing 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDAC or EDC), sulfo-N-hydroxysuccinimide (Sulfo-NHS) and ethanolamine-HCl
- Preconditioning Reagents (see Recipes)
- Immobilization Buffer (see Recipes)
- Running Buffer (see Recipes)
- Running Buffer with 5% DMSO (see Recipes)
Equipment
- ProteOn Standard & Deep-Well Microplates (Bio-Rad Laboratories, catalog numbers: 176-6020 and 176-6023 )
- ProteOn Microplate Sealing Film (Bio-Rad Laboratories, catalog number: 176-6040 )
- Nalgene Rapid-Flow Filter Units and Bottle Top Filters, PES Membrane, Sterile (VWR International, catalog number: 16211-056 ) (for the 1 L size)
- ProteOn XPR36 instrument (Bio-Rad Laboratories)
- BenchTop Centrifuge for microfuge tubes
- BenchTop Centrifuge to spin ProteOn 96-well assay plate
Software
- ProteOn Manager Software Version 3.1.0.6
- Scrubber software designed for ProteOn data analysis (BioLogic Software)
Procedure
- Prime the system with distilled water. Precondition a GLH chip using 1-min pulses each of 100 mM HCl, 50 mM NaOH, 0.5% SDS and 10% DMSO using a flow rate of 30 µl/min in both the horizontal and vertical directions. Seal all microplates used with ProteOn sealing film.
- Prime the system with Running Buffer. Activate all 6 ligand channels (L1-L6) in the vertical direction using EDC and sulfo-NHS as prescribed by the manufacturer using a flow rate of 30 µl/min and a contact time of 5 min.
- Dilute NS5B protein in Immobilization Buffer to a concentration of 30 µg/ml and inject using a flow rate of 30 µl/min for 5 min in the vertical direction into the ligand channel(s) to obtain 5,000-10,000 RU of immobilized protein (see Notes). Leave the L1 channel open as a reference channel.
- When calculating the theoretical maximum response, Rmax, the activity of the protein should be taken into account. Rmax is calculated as RL * (MWA/MWL) * n, where RL is equal to the amount of ligand immobilized, MWA is the molecular weight of the analyte, MWL is the molecular weight of the ligand, and n is the number of binding sites per ligand molecule. Assuming single-site binding (i.e. n = 1), a small molecule analyte having an average molecular weight of 500 Da interacting with a surface comprised of 10,000 RU of NS5B ΔC21 (M.W.~ 64,300 Da) should yield a theoretical Rmax of ~ 78 RU (i.e. 10,000 RU x 500 Da/64,300 Da) or 39 RU if the surface is only 50% active.
- Deactivate all vertical channels (L1-L6) by injecting ethanolamine-HCl for 5 min at flow rate of 30 µl/min.
- Rotate the multi-channel module (MCM) as all subsequent injections will occur in the horizontal direction. Prime the system with Running Buffer containing 5% DMSO and also use this buffer to inject buffer three times to help stabilize the system after rotation.
- To account for excluded volume effects, a set of five solutions between 4.5% and 5.5% DMSO (DMSO concentration series) are injected for the software to generate a DMSO excluded volume calibration (EVC) curve. Prepare a 5.5% DMSO solution by adding 15 µl of 100% DMSO to 3,000 µl of Running Buffer with 5% DMSO, and a 4.5% DMSO solution by adding 333 µl of Running Buffer to 3,000 µl of Running Buffer with 5% DMSO. Mix the 5.5% and 4.5% DMSO solutions according to the volumes prescribed below to obtain the five different solutions. Five separate injections are performed to generate a DMSO concentration series each corresponding to the injection of one of the DMSO solutions. The software will use this series of injections to generate a DMSO calibration curve to correct for excluded volume effects.
5.5% DMSO buffer 4.5% DMSO buffer
1200 µl 0 µl
900 µl 300 µl
600 µl 600 µl
300 µl 900 µl
0 µl 1200 µl - When designing the analyte injection sequence, include buffer injections between every injection of compound (or as space permits in the 96-well plate).
- Thaw compounds, typically stored in 100% DMSO in microfuge tubes at -20 °C, and quickly spin them in a microfuge centrifuge.
- Serially dilute the compounds in DMSO to achieve a set of five concentrations (four-fold dilutions in this example) at 20 times the desired final assay concentration. Transfer the diluents to wells in the microplate containing an appropriate volume of Running Buffer to yield a final DMSO concentration of 5% (i.e. matching the % of DMSO in Running Buffer with DMSO). These will be injected as Analytes A2-A6. Add 100% DMSO to the same volume of Running Buffer to the well corresponding to injection of Analyte A1 to yield a final DMSO concentration of 5%. Injection from well A1 will serve as a blank and be used to double-reference the data.
- Centrifuge the plate using a quick 15-30 second pulse to ensure collection of any compound solution stuck to the walls of the microplate well.
- Inject compounds at 100 µl/min for a contact time of 90 sec and for variable dissociation times ranging between 60 sec and one hour (as required by individual compounds). Absent any knowledge about a compound’s affinity, preliminary experiments involving injection at a single concentration may have to be performed in order to gauge optimal dissociation times for each compound.
- Data can be analyzed using ProteOn Software but can also be analyzed using a version of Scrubber software designed for ProteOn data. Double reference all signals (using L1 & A1) and correct for excluded volume effects by applying the DMSO calibration curve.
- Sensorgrams for all the concentrations tested should be fit simultaneously, i.e. globally. In general, data should be always first fit to a simple 1:1 kinetic model which yields the equilibrium dissociation constant (KD), the maximum binding capacitance of the surface Rmax, (which may differ from the theoretical maximum binding capacitance) and the rate constants of association (ka) and dissociation (kd). In a simple 1:1 kinetic model, single values of ka, kd and Rmax are obtained. Both Scrubber and ProteOn fitting routines solve for the parameters ka and kd. KD is then calculated as the ratio of kd/ka. Use of other more complex models that require more than one value for any of these parameters should be explicitly stated. Filibuvir binding to NS5B ΔC21 (see Figure 1) is an example of a small molecule binding showing simple 1:1 kinetics.
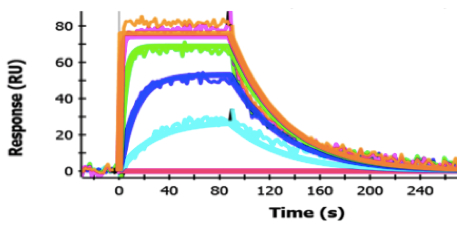
Figure 1. Filibuvir Binding to NS5B ΔC21. Sensorgrams of Filibuvir binding are shown. Filibuvir was injected for a contact time of 90 sec at concentrations of (from top to bottom) 4 µM (orange), 1 µM (pink), 0.25 µM (green), 0.0625 µM (blue) and 0.0156 µM (cyan). Dissociation was monitored for 180 sec. The fit to the data are shown as solid lines.
Notes
The physiological immobilization buffer used can readily pre-concentrate sufficient NS5B enzyme onto the carboxyl surface. The ligand activity increased from 45% to 99% for NS5B ΔC21 when opting to dilute protein in HEPES-buffered saline rather than standard low pH (5.5) 10 mM sodium acetate immobilization buffer.
Recipes
- Preconditioning Reagents
100 mM HCl
50 mM NaOH
0.5% SDS
10% DMSO - Immobilization Buffer
10 mM HEPES (pH 7.5)
150 mM NaCl
Filter sterilize (0.45 μm) - Running Buffer
50 mM HEPES
5 mM MgCl2
10 mM KCl
1 mM EDTA
1 mM TCEP
0.01% P20
Adjust pH to 7.5 with NaOH
Add dH2O to the desired volume
Filter sterilize (0.45 μm) - Running Buffer with 5% DMSO
Add 100% DMSO to Running Buffer to a final concentration of 5% DMSO
For example, add 50 ml of DMSO to Running Buffer to a total volume of 1 L.
Acknowledgments
A brief description of this protocol has previously appeared in Boyce et al. (2014).
References
- Boyce, S. E., Tirunagari, N., Niedziela-Majka, A., Perry, J., Wong, M., Kan, E., Lagpacan, L., Barauskas, O., Hung, M., Fenaux, M., Appleby, T., Watkins, W. J., Schmitz, U. and Sakowicz, R. (2014). Structural and regulatory elements of HCV NS5B polymerase - beta-loop and C-terminal tail - are required for activity of allosteric thumb site II inhibitors. PLoS One 9(1): e84808.
- Hung, M., Wang, R. and Liu, X. (2011). Preparation of HCV NS3 and NS5B proteins to support small-molecule drug discovery. Curr Protoc Pharmacol Chapter 13: Unit13B 16.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wong, M. and Papalia, G. A. (2014). A Surface Plasmon Resonance Method to Study HCV NS5B Inhibitors . Bio-protocol 4(4): e1044. DOI: 10.21769/BioProtoc.1044.
Category
Biochemistry > Protein > Interaction > Protein-ligand interaction
Microbiology > Microbial biochemistry > Protein > Interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










