- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Dissociated Retinal Cell Culture
Published: Vol 4, Iss 2, Jan 20, 2014 DOI: 10.21769/BioProtoc.1033 Views: 16559
Reviewed by: Xuecai Ge

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2243 Views

Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System
Wei Tan [...] Qing Li
Nov 20, 2025 1343 Views

Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
Jianwei Li [...] Guohui Lu
Dec 5, 2025 1475 Views
Abstract
The retina is a relatively simple and accessible part of the central nervous system, making it a powerful model to study cell fate specification mechanisms. Multipotent retinal progenitor cells (RPCs) give rise to seven major classes of retinal cell types. Mechanisms regulating cell fate choice in the retina depend on both cell intrinsic and environmental factors, but their relative contribution to specific cell fate decisions remains unclear. Dissociated retinal cell cultures provide a great assay to study this problem. RPCs are cultured in serum-free and extract-free medium, providing the investigator with a control over the environment to address questions related to the effects of a particular molecule on the development of retinal neurons. In addition, dissociated cell cultures can be used to study the importance of cell intrinsic mechanisms by isolating RPCs from their normal environment (Cayouette et al., 2003; Jensen and Raff, 1997). The method described below is suitable for the clonal-density culture of RPCs. In such cultures, RPCs are isolated from each other and from the postmitotic neurons. They divide and differentiate into different retinal cell types to form small colonies, or “clones”. In a recent study, we found that these clones are indistinguishable from the clones that develop in situ in the retina, both in terms of cell number and cell type composition, suggesting that intracellular mechanisms play a key role in retinal development (Cayouette et al., 2003).
Keywords: RetinaMaterials and Reagents
- Animals
We typically use retinas from albino Sprague Dawley rats. This method works for culturing rat retinas aged between embryonic day 17 to post-natal day 1. Younger and older retinal cells or retinal cells from mouse retinas do not seem to survive as well under these conditions, but can also be cultured. - DPBS (Life Technologies, catalog number: 14040 )
- Neurobasal medium (Life Technologies, catalog number: 21103-049 )
- DMEM/F12 medium (Life Technologies, catalog number: 10565-018 )
- B27 supplement (Life Technologies, catalog number: 17504-044 )
- Pen/Strep (P/S) (Life Technologies, catalog number: 15140-122 )
- Poly L-lysine (PLL) (Sigma-Aldrich, catalog number: P-4707 )
- Mouse Laminin (Life Technologies, catalog number: 23017-015 )
- Papain (Worthington Biochemical, catalog number: LS003126 )
- L-cystein crystal (Sigma-Aldrich, catalog number: C-7352 )
- Trypsin Inhibitor (Roche Diagnostics, catalog number: 109878 )
- 0.45 µm and 0.22 µm filters (Fisher Scientific, catalog number: SLHV033RS and SLGP033RS )
- Hoechst 33342 (Life Technologies, Molecular Probes®, catalog number : H3570 )
- “Incomplete” RGM medium
- Growth factors (see Recipes)
- 10x Lo Ovomucoid (see Recipes)
- 20x 4% BSA (Sigma-Aldrich, catalog number: A-4161 ) (see Recipes)
- 0.4% DNase (Worthington Biochemical, catalog number: LS002007 ) (see Recipes)
- Insulin (2.5 mg/ml) (Sigma-Aldrich, catalog number: I-6634 ) (see Recipes)
- cpt-cAMP (Sigma-Aldrich, catalog number: C-3912 ) (see Recipes)
- N-acetyl cysteine (NAC) (Sigma-Aldrich, catalog number: A-9165 ) (see Recipes)
- Forskolin (Sigma-Aldrich, catalog number: D-2438 ) (see Recipes)
- N2 Supplement (see Recipes)
- 4% Paraformaldehyde (Electron Microscopy Science, catalog number: 15710 ) (see Recipes)
- Apo-Transferrin (Sigma-Aldrich, catalog number: T-1147 ) (see Recipes)
- Progesterone (Sigma-Aldrich, catalog number: P-8783 ) (see Recipes)
- Putrescine (Sigma-Aldrich, catalog number: P-5780 ) (see Recipes)
- Sodium Selinite (Sigma-Aldrich, catalog number: S-5261 ) (see Recipes)
Equipment
- 35 mm petri dish (BD Biosciences, Falcon®, catalog number: 353001 )
- 50 ml conical tube
- CO2 incubator
- 15 ml conical tube
- 100 mm petri dish
- CO2 chamber
- Straight forcep
- Fine forcep (Fine Science Tools, catalog number: 11252-23 )
- Curved forcep (Fine Science Tools, catalog number: 91197-00 )
- Sharp scissor
- Laminar flow hood
- Water bath
- P1000 pipette
- Microscope
- Hemacytometer
- Centrifuge
Procedure
Day before experiment
- Coat culture dishes with poly-L-lysine and laminin
- Prepare a 50% working solution of Poly L-lysine (PLL) by diluting the stock solution in sterile water. Add 1.5 ml of PLL solution to each 35 mm petri dish. Leave at RT for 60 min. Remove PLL and let dry in the hood. Wash three times with sterile water.
- Prepare the mouse laminin solution at a concentration of 5 µg/ml in cold Neurobasal medium. Add 1.5 ml of mouse laminin solution to the dish.
- Incubate for at least overnight in an incubator equilibrated at 37 °C and 8% CO2. 5% CO2 incubation is ok, but not optimal for the Neurobasal/DMEM/F12 medium mix. Longer incubation can be done and were found to improve quality of the prep.
- Prepare a 50% working solution of Poly L-lysine (PLL) by diluting the stock solution in sterile water. Add 1.5 ml of PLL solution to each 35 mm petri dish. Leave at RT for 60 min. Remove PLL and let dry in the hood. Wash three times with sterile water.
- Prepare the “incomplete” retina growth medium (RGM)
- In a 50 ml conical tube, under sterile condition, add to following:
10 ml of Neurobasal medium
10 ml of DMEM/F12 medium
100 µl of N2 supplement
200 µl of B27 supplement
200 µl of 100x Pen/Strep (P/S)
200 µl of 6 mg/ml NAC
40 µl of 50 mM cpt-cAMP
20 µl of 10 mM Forskolin
200 µl of 2.5 mg/ml Insulin
Filter sterilize the solution using a 0.22 µm filter previously rinsed with Neurobasal medium and stored at 4 °C until needed.
- In a 50 ml conical tube, under sterile condition, add to following:
Day of Experiment
- Prepare the solution needed (see Recipes for the preparation of the stock solutions)
- “Incomplete” RGM medium add the growth factors
4 µl of EGF 250 µg/ml
2 µl of b-FGF 100 µg/ml
20 µl of BDNF 50 µg/ml
5 µl of NT-3 100 µg/ml
To equilibrate the medium at the right temperature and pH, put the lid on the 50 ml tube and unscrew ¼ of a turn. Place the tube in the incubator at 37 °C and 8% CO2 until needed. - Papain solution
In a 15 ml conical tube add the following
10 ml of DPBS
5 µl of 1 N NaOH
100 U of papain solution
100 µl of 0.4% DNase
2 mg of L-cystein crystal (to be added only prior to using the solution for dissociation)
Keep at 37 °C in a water bath - LO-OVO solution
9 ml of DPBS
1 ml of 10x LO-OVO
100 µl of 0.4% DNase
Keep at 37 °C in a water bath
- “Incomplete” RGM medium add the growth factors
- Culture procedure
Note: The dissection should be carried out under sterile conditions. Spray the working area with 70% ethanol and use a flame. Sterilize all the instruments before use.
Prepare two 100 mm dish filled with 10 ml of sterile DPBS, one to transfer the heads and the other to dissect the eyes and two 35 mm dish filled with 2 ml of sterile DPBS to collect the retinas.- Collection of the embryos
- Euthanize the animal in a CO2 chamber.
- Place the animal on it’s back and clean the belly skin with 70% ethanol.
- Pinch the skin with a straight forcep, lift the skin and muscle and make a V shape opening using scissors on both side of the abdomen starting from the lower abdomen and cutting all the way to the side of the animal. Lift the skin and muscle upward to reveal the internal organs and uterus.
- Cut out the uterus and transfer it to a 150 mm dish.
- Remove the embryos from the uterus and decapitate the heads with sharp scissors.
- Transfer the heads to a 100 mm dish filled with DPBS.
- Euthanize the animal in a CO2 chamber.
- Dissection of the retinas
- Carefully remove the skin that covers the eye.
- Place a curved forcep of both side of the eye and apply a gentle pressure. The eye should pup out.
- Pinch the back of the eye and pull.
- Transfer the eye in a 100 mm dish filled with DPBS.
- With the lens facing downward, secure the eye in position with a fine forceps and remove the optic nerve (Figure 1, steps 1-2).
- Introduce the tip of another forceps through the optic nerve head and between the choroid and retina to secure the sclera.
- Now that the sclera is secured, you need to gently tear it open with both forceps from the optic nerve head to the cornea to expose the retina andlens (Figure 1, steps 3-4).
- Carefully detach the lens and place the retinas in a 35 mm dish filled with DPBS (Figure 1, steps 5-6). From E18-19 onward, it is required to remove the blood vessel covering the retina to avoid endothelial cell contamination in the culture.
- The blood vessels form a reddish membrane overlying the inner face of the retina. The membrane is easily removed by pinchingand pulling with a fine forceps.
- Transfer the retinas in a 35mm dish filled with DPBS. Using a fine scalpel, slice the retinas in small pieces (2 mm) to help the dissociation. The number of retinas to dissect depends on your need. Typically, the yield is about 0.5 x 106 cells for one E17 retina and 1.6 x 106 cells for one P0 retina. It is optimal to dissect at least 6 to 8 retina as it improve cell recovery.
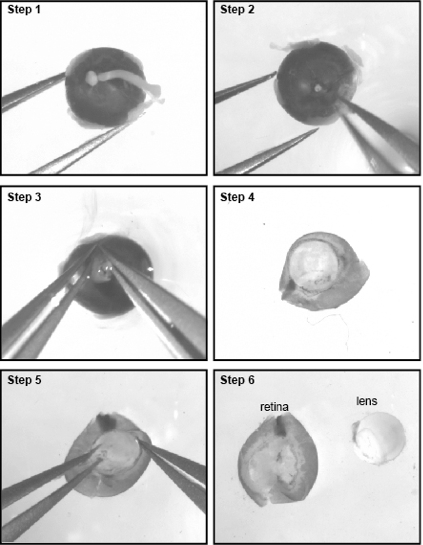
- With the lens facing downward, secure the eye in position with a fine forceps and remove the optic nerve (Figure 1, steps 1-2).
- Carefully remove the skin that covers the eye.
- Dissociation of the retinas (all these steps are done in the laminar flow hood)
- Digestion of the tissue
Take out the papain tube from the water bath and add 2 mg of L-cystein.
Transfer the pieces of retina in this tube.
Return the tube in the 37 °C water bath and incubate 5 min if you are working with E17 retinas or 8 min if you are working with P0 retinas. - Neutralisation of the enzyme activity
Take out the tube from the water bath, let the pieces of retina settle in the bottom of the tube and remove the supernatant with gentle aspiration.
Slowly add 4 ml of LO-OVO solution over the pieces of retina.
Let the pieces of retina settle in the bottom of the tube and remove the supernatant with gentle aspiration. Add 1 ml of LO-OVO solution. - Trituration
Using the P1000, gently triturate up and down the pieces of retina to break them apart until the solution appears cloudy.
Make sure that the cell suspension is well dissociated and that no clumps of cells remain by observing a small aliquot of the suspension under the microscope.
If clumping is observed, triturate the suspension with a little more force until unicellular suspension is obtained. (If the cells are difficult to dissociate, the incubation time in papain can be increased). Be careful not to over triturate the cells, this could lead to decreased survival.
Add the remaining 5 ml of LO-OVO solution to the cell suspension and centrifuge 11 min in a benchtop centrifuge at 200 x g at room temperature.
- Digestion of the tissue
- Plating
- Take out the RGM medium from the incubator.
- Aspirate the supernatant and gently resuspend the cell pellet in 1 ml of equilibrated RGM medium.
- Using the hemacytometer, count the number of total cells. For clonal density culture, add 5,000 cells per 35 mm dish, for low density culture, add 20,000 cells per 35 mm dish and for high density culture, add 100,000 cells per 35 mm dish.
- Dilute to right amount of cells in 1 ml of RGM medium and add it to the 35 mm dish.
- Place in the incubator at 37 °C with 8% CO2 for 2 h.
- Once the cells have adhered to the bottom of the dish, gently add another 1.5 ml of medium and replace in the incubator.
- Take out the RGM medium from the incubator.
- Feeding
Feed the cells every 3 to 4 days by removing 50% of the culture medium and adding the same amount of equilibrated RGM medium. In optimal conditions, the cells can be cultured for 10 to 14 days.
- Collection of the embryos
- Immunostaining of cultured cells
Note: In these culture conditions, retinal progenitor cells will divide and differentiate into the different retinal cell types, normally produced from E17 (amacrines, bipolars, rod photoreceptors, and Müller cells). Various cell-specific markers can be used to identify the different retinal cell types. Nestin for retinal progenitor cells, Brn3b and neurofilamment 165 for ganglion cells, Rhodopsin and recoverin for rod photoreceptors, Islet-1 for bipolar cells and a subtype of amacrines and ganglion cells, whereas Pax-6 is a marker of retinal progenitor cells, amacrines and ganglion cells. Here, we provide a general protocol for immunostaining.- Aspirate the culture medium and fix the cells by adding cold 4% PFA for 15 min on ice.
- Rinse three times with 1x PBS.
- Block/permeabilize for 1 h at room temperature (RT) with block/permeabilization solution (20% goat serum + 0.4% Triton in 1x PBS).
- Prepare the primary antibody in 1 ml of block/permeabilization solution. Dilution to be used depend on the antibody.
- Incubate overnight at 4 °C.
- Discard primary antibody.
- Rinse three times with 1x PBS.
- Prepare the secondary antibody in 1 ml of block/permeabilization solution. The secondary antibody is usually used at a 1:1,000 dilution.
- Incubate 1 h at RT.
- Discard secondary antibody.
- Rinse three times with 1x PBS.
- To counterstain the nucleus, incubate in Hoechst solution (1 ml of Hoechst in 10 ml of 1x PBS) for 5 min at RT.
- Observe under inverted microscope.
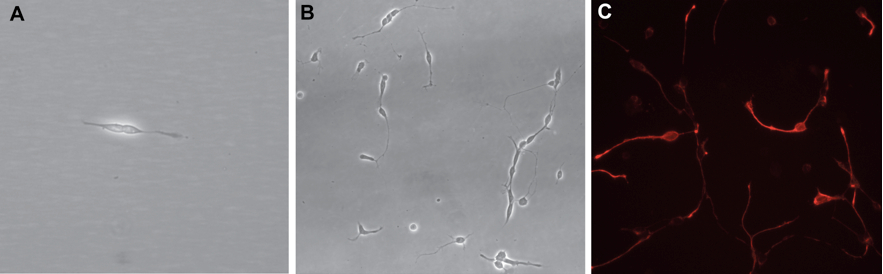
Figure 2. Phase contrast and immunostaining micrographs of dissociated cultures. A. Two retinal progenitor cells 18 h after plating. Retinal progenitors have a typical bipolar morphology. B. A dissociated retinal cell culture seen after 7 days in culture showing many differentiated neurons. C. Retinal progenitor cells stained for Nestin (red) 24 h after plating.
- Aspirate the culture medium and fix the cells by adding cold 4% PFA for 15 min on ice.
Recipes
- Growth factors
- b-FGF: Reconstitute in 10 mM Tris pH 8.5 to a concentration of 100 µg/ml
- EGF: Reconstitute in water to a concentration of 250 µg/ml
- NT-3: Reconstitute in water to a concentration of 100 µg/ml
- BDNF: Dilute 1mg/ml stock solution to a concentration of 50 µg/ml in water
- b-FGF: Reconstitute in 10 mM Tris pH 8.5 to a concentration of 100 µg/ml
- 10x Lo Ovomucoid
To 40 ml DPBS add 600 mg BSA, Mix well
Add 600 mg Trypsin inhibitor and mix to dissolve
Adjust pH to 7.4; requires the addition of approx. 400 µl of 1 N NaOH
When completely dissolved, filter through a 0.22 µm filter pre-rinsed with PBS
Make 1.0 ml aliquots and store at -20 °C - 20x 4% BSA
Add 2 g BSA in 50 ml DPBS
Dissolve at 37 °C. Adjust pH to 7.4 (with approximately 200 µl 1 N NaOH)
Filter sequentially through 0.45 µm and 0.22 µm filters, pre-rinsed with PBS
Make 1.0 ml aliquots
Stored at -20 °C - 0.4% DNase
Dissolve 40 mg in 10 ml of sterile water
Mix by gentle inversion
Aliquots in 200 μl and stored at -20 °C - Insulin (2.5 mg/ml)
To 10 ml sterile water add
25 mg insulin
50 µl 1.0 N HCl
Mix well
Stored at 4 °C for no more than 4 to 6 weeks - cpt-cAMP (50 mM)
Dissolve the cpt-cAMP in sterile water at 25 mg/ml
Aliquots in 200 µl and stored at -20 °C - NAC (6 mg/ml)
Dissolve 6 mg N-acetyl cysteine in 1 ml of Neurobasal medium
Aliquots in 200 µl
Stored at -20 °C - Forskolin (10 mM)
Add 2.4 ml DMSO to a 10 mg bottle (4.2 mg/ml or 10 mM)
Make 50 µl aliquots
Stored -20 °C - N2 Supplement
- Prepare N2 stock solutions
- Apo-Transferrin: Dissolve 500 mg in 5 ml of water (100 mg/ml stock solution), filter (0.22 µM), aliquot and stored at -20 °C
- Progesterone: Dissolve 6 mg in 10 ml of ethanol to make 0.6 mg/ml stock solution, filter (0.22 µM), aliquot and stored at -20 °C
- Putrescine: Dissolve 1.6 g in 10 ml of water to make 1 M (160 mg/ml) stock solution, filter (0.22 µM), aliquot and stored at -20 °C
- Sodium Selenite: Dissolve 2.59 mg in 5 ml of water to make 3mM stock solution, filter (0.22 µM), aliquot and stored at -20 °C
- BSA: Dissolve 500 mg in 10 ml of water to make 50 mg/ml stock solution, filter (0.22 µM), aliquot and stored at -20 °C
- Apo-Transferrin: Dissolve 500 mg in 5 ml of water (100 mg/ml stock solution), filter (0.22 µM), aliquot and stored at -20 °C
- Prepare 10 ml of 100x N2 solution
To 7.857ml of DMEM-F12, add the following:
1 ml of Apo-Transferrin 100 mg/ml
33 µl of progesterone 0.6 mg/ml
100 µl of putrescine 1 M
10 µl of sodium selenite 3 mM
1 ml of BSA 50 mg/ml
Aliquot in 200 µl and stored at -20 °C
- Prepare N2 stock solutions
- 4% Paraformaldehyde
Dilute a vial of 16% PFA (10 ml) in 26 ml of water + 4 ml of 10x PBS; This solution can be kept at 4 ˚C for no longer than 1 month.
Acknowledgments
Funding for this work was provided by the Canadian Institutes of Health Research and the Foundation Fighting Blindness Canada. This protocol was adapted from procedures published in Cayouette et al. (2003). The authors wish to thank members of the Cayouette lab, past and present, for continuous support and improvements on the protocol over the years.
References
- Cayouette, M., Barres, B. A. and Raff, M. (2003). Importance of intrinsic mechanisms in cell fate decisions in the developing rat retina. Neuron 40(5): 897-904.
- Jensen, A. M. and Raff, M. C. (1997). Continuous observation of multipotential retinal progenitor cells in clonal density culture. Dev Biol 188(2): 267-279.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Jolicoeur, C. and Cayouette, M. (2014). Dissociated Retinal Cell Culture. Bio-protocol 4(2): e1033. DOI: 10.21769/BioProtoc.1033.
- Kechad, A., Jolicoeur, C., Tufford, A., Mattar, P., Chow, R. W. Y., Harris, W. A. and Cayouette, M. (2012). Numb is required for the production of terminal asymmetric cell divisions in the developing mouse retina. J Neurosci 32(48): 17197-17210.
Category
Neuroscience > Development > Retinal culture
Cell Biology > Cell isolation and culture > Cell isolation
Cell Biology > Tissue analysis > Tissue isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link