- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Membrane Preparation, Sucrose Density Gradients and Two-phase Separation Fractionation from Five-day-old Arabidopsis seedlings
Published: Vol 3, Iss 24, Dec 20, 2013 DOI: 10.21769/BioProtoc.1014 Views: 26897
Reviewed by: Tie Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Streamlining Protein Fractional Synthesis Rates Using SP3 Beads and Stable Isotope Mass Spectrometry: A Case Study on the Plant Ribosome
Dione Gentry-Torfer [...] Federico Martinez-Seidel
May 5, 2024 2869 Views

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1989 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1772 Views
Abstract
Membrane preparation has been widely used for characterization the membrane proteins. Membrane fractions can be separated by a combination of differential and density-gradient centrifugation techniques (Hodges et al., 1972; Leonard and Vanderwoude, 1976). Here we firstly describe a method to isolate total microsomal fractions including plasma membrane, intracellular vesicles, Golgi membranes, endoplasma reticulum, and tonoplast (vacuolar membrane) from 5-7 days old seedlings, which is often analyzed for auxin transporters in Arabidopsis (Leonard and Vanderwoude, 1976; Titapiwatanakun, et al., 2009; Yang et al., 2013; Blakeslee et al., 2007). After homogenization, plant debris including cell walls, chloroplasts and nucleus were removed by low speed centrifugation (8,000 x g), then total microsomal membranes were pelleted by high speed centrifugation (10,000 x g) and separated from soluble fractions. We secondly describe a method to separate microsomal fractions according to size or density in a sucrose density-gradient system by centrifugation. The linear sucrose gradient from 20%-55% (1.09-1.26 g cm-3) were used to separate membranes with different densities: tonoplast, 1.10-1.12 cm-3, Golgi membranes, 1.12-1.15 cm-3, rough endoplasmic reticulum 1.15-1.17 cm-3, thylakoids, 1.16-1.18 cm-3, plasma membrane, 1.14-1.17 g cm-3, and mitochondrial membranes, 1.18-1.20 cm-3 (Leonard and Vanderwoude, 1976; Larsson et al., 1987; Briskin and Leonard, 1980). However, the plasma membrane can also be isolated according to its outer surface properties which are very different from intracellular membrane surfaces. Thus, the right-side-out plasma membrane vesicles can be separated in an aqueous Dextran-polyethylene glycol two-phase system. The plasma membranes can be purified to > 90% in the upper phase (Larsson et al., 1987; Alexandersson et al., 2008). Two-phase systems for Arabidopsis seedlings were described in the section 3. Sucrose density gradient membrane fractionation followed by western blot is often used to analyze the distribution of certain membrane protein, while Two-phase separation is used when high purity of plasma membrane or intracellular membrane is required.
Materials and Reagents
Note: All chemicals were purchased from Sigma-Aldrich (http://www.sigmaaldrich.com/united-states.html) unless otherwise specified.
- Arabidopsis seedlings or mature tissue
- Ice
- Sucrose (Molecular Biology, Sigma-Aldrich, catalog number: 84097 )
- HEPES (Sigma-Aldrich, catalog number: H-3375 )
- EDTA (Research Products International, catalog number: E57020 )
- PVP (40,000) (Fisher Scientific, catalog number: BP431 )
- BSA (Sigma-Aldrich, catalog number: A-7906 )
- DTT (Sigma-Aldrich, catalog number: D0632 )
- Leupeptin (Sigma-Aldrich, catalog number: L2884 )
- PMSF (Sigma-Aldrich, catalog number: P-7626 )
- Benzamidine (Sigma-Aldrich, catalog number: B-6506 )
- Pepstatin A (Sigma-Aldrich, catalog number: P5318 )
- Aprotinin (Sigma-Aldrich, catalog number: A1153 )
- BTP-MES (BIS-TRIS propane, Sigma-Aldrich, catalog number: B6755 ; MES, Research Products International, catalog number: M22040 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- Dextran (GE, catalog number: 17-0320-01 )
- Polyethylene glycol 3350 (Union Carbide Corporation, catalog number: Carbowax 3350 )
- EGTA (Sigma-Aldrich, catalog number: E3889 )
- Protease inhibitor cocktail (Sigma-Aldrich, catalog number: P8340 )
- Grinding buffer (see Recipes)
- Resuspension buffer (see Recipes)
- Stock solutions (see Recipes)
- Phase mixture (see Recipes)
- Phase system (see Recipes)
Equipment
- Hermle Labnet Z383 centrifuge (Labnet International)
- OptimaTM-L-90K ultracentrifuge (Beckman Coulter)
- TLX centrifuge (TLA 100.3 rotor) (Beckman Coulter)
- Waring blender jar (Waring Pro model: PBB212 )
- Mortar and pestle
- SW-28 rotor
- Nylon SS-34 tube
- SW-38 tube
- 3.5 ml polycarbonate TLA 100.3 tube
- 2 ml screwcap tubes
- 10, 15 and 50 ml Falcon tubes
- Polyallomer ultracentrifuge tube (Beckman Coulter)
Procedure
- Membrane preparation
- Turn on Hermle Labnet Z383 centrifuge. Set temperature to 4 °C. Collect 3-5 g of tissue from 5-d seedlings or mature tissue. Squeeze balls of tissue dry and weigh.
- For large quantity samples (> 40 g), add tissue to Waring blender jar and fill jar so that no air is present. Fill a bucket with ice. Pulse the waring jar for 30 s, then incubate in ice for 5 min. Repeat 3x.
- For small quantity samples (< 10 g), this step can be skipped, and the seedlings can be ground in grinding buffer in a mortar and pestle (see step A2).
- For large quantity samples (> 40 g), add tissue to Waring blender jar and fill jar so that no air is present. Fill a bucket with ice. Pulse the waring jar for 30 s, then incubate in ice for 5 min. Repeat 3x.
- Pour into pre-chilled motar and pestle on ice. Add a little bit of sand (acid washed, Sigma 18649).
- Immediately grind in a mortar and pestle for approximately 10 min.
- Pour through Miracloth lining a plastic power funnel set into nylon SS-34 tube. Squeeze through Miracloth to extract all of liquid homogenate. Fill tubes to 1 cm from top when standing upright.
- If the yield is poor, or the sample still look “unground”, it can be returned to mortar and pestle, and ground again in a small aliquot of grinding buffer.
- Immediately grind in a mortar and pestle for approximately 10 min.
- Balance the tubes and centrifuge at 8,000 x g, 4 °C for 15 min.
- Turn on Beckman Coulter OptimaTM-L-90K ultracentrifuge and set temperature to 4 °C.
- Carefully pour off supernatant from SS-34 tubes immediately without disturbing the pellet. Try not to collect any of the dark green pellet material in the supernatant fraction (the pellet is very easy to disturb).
- Collect the cellular debris pellet and store in liquid nitrogen until use, if desired.
- Turn on Beckman Coulter OptimaTM-L-90K ultracentrifuge and set temperature to 4 °C.
- To obtain a microsomal pellet, centrifuge the supernatant from the 8,000 x g spin at 100,000 x g, 4 °C, for 50 min, 1 h. Store additional supernatant on ice.
- Use thin wall tubes in the SW-28 rotor. The entire bucket assembly must be balanced within 0.005 g. Be sure to place buckets in the correct position and verify that they have engaged the rotor properly.
- Pour off supernatant and store at -70 °C (if soluble proteins are needed).
- If you have additional supernatant (from the 8,000 x g spin) on ice, pour off enough supernatant (from the 100,000 x g spin) from the ultracentrifuge tubes to allow for refilling with extra 8,000 x g supernatant. Balance tubes with poured off 100,000 x g supernatant as needed. Repeat 50 min spin.
- Use thin wall tubes in the SW-28 rotor. The entire bucket assembly must be balanced within 0.005 g. Be sure to place buckets in the correct position and verify that they have engaged the rotor properly.
- Pour off all supernatants–save as above if desired. Wipe out inside of tubes above the pellet with a Kimwipe or paper towel. Place on ice.
- Re-suspend each SW-28 pellet in microsomal resuspension buffer (volume will depend on pellet size and intended use, usually 1-2 ml). Use a cut-off 1 ml pipet tip to resuspend initially. This will be difficult, as the microsomes are very stick and hydrophobic. Try not to let the pellet stick to the pipet tip or walls of the centrifuge tube.
- After the pellet has been fairly well resuspended (pipet up and down for 100 times), use a normal 1 ml pipet tip to “break up” the clumps a bit and resuspend the microsomes more thoroughly (pipet up and down for 100 times).
- Finally, use a 200 μl pipet tip to break up the clumps and completely homogenize the preparation (pipet up and down for 200 times).
- If a wash of the pellet is desired, transfer the resuspended pellet to 3.5 ml polycarbonate TLA 100.3 tube. Wash each SW-38 tube with a bit of additional buffer, and add this to the TLA tubes. Centrifuge at 100,000 x g, 4 °C for 30 min in the TLX centrifuge (TLA 100.3 rotor). Discard supernatant, wipe out walls of tubes, store on ice.
- After the pellet has been fairly well resuspended (pipet up and down for 100 times), use a normal 1 ml pipet tip to “break up” the clumps a bit and resuspend the microsomes more thoroughly (pipet up and down for 100 times).
- Resuspend the microsomal pellet in resuspension buffer as in step A6. If fraction is being stored, aliquot into 500 μl–1 ml traction in 2 ml screwcap tubes. Freeze in liquid nitrogen, and store in either liquid nitrogen or -80 °C freezer until use.
- Turn on Hermle Labnet Z383 centrifuge. Set temperature to 4 °C. Collect 3-5 g of tissue from 5-d seedlings or mature tissue. Squeeze balls of tissue dry and weigh.
- Sucrose density gradients
Note: Preparation of sucrose solution: Important! Sucrose must be ultra pure.- Prepare 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55% (w/v) sucrose solution with 20 mM HEPES pH= 7.8, 1 mM EDTA in 10 ml tubes.
Note: Turn the tubes on a rotator for approximately 20 min until all sucrose has dissolved. - On the bottom of tube (Beckman polyallomer ultracentrifuge tube) put first the drop of 55% Suc and after that 2 ml of 50%, 45%, 40%, 35%, 30%, 25% and 20% were carefully added on the top, the interface should be seen (see Figure 1). At the top of the tube upon which to pipette the 0.5 ml sample of resuspended microsomal fraction (from membrane preparation).
Note: Allow sucrose to run down very slowly inside of tube. Sucrose layers should be visible when they are layering progressively. Place the tube with sucrose layers at 4 °C overnight to form continuous gradients.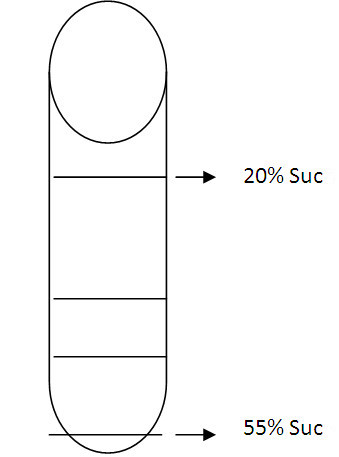
Figure 1. A carton to show the interface of sucrose layers. All interfaces should be clearly seen between different concentrations. Please carefully add each concentration to obtain clear interface. - Centrifuge (Optima L-90K ultracentrifuge) 100,000 x g for 12 h.
- Immediately after the run the tube should be removed from the rotor taking care not to disturb the layers of sucrose gradient.
- Collect the 0.5 ml fractions from top to bottom very carefully to Eppendorf’s tubes.
- Prepare 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55% (w/v) sucrose solution with 20 mM HEPES pH= 7.8, 1 mM EDTA in 10 ml tubes.
- Two phase separation for plasma membrane and endomembrane fractions
Stock solutions, Phase mixture (2.7 g) and Phase system (30 g) (see Recipes)- Microsomal membranes are prepared as section A above.
- Fresh microsomal membranes are resuspended in 1 ml of buffer (0.33 M sucrose, 3 mM KCl, and 5 mM potassium phosphate, pH 7.8, and 20 mg/ml complete protease inhibitor cocktail from Sigma-Aldrich).
- Add 0.9 g resuspended membrane to 2.7 g two-phase separation mixture (can be scaled up).
- After mixing (inverting 30 times), the phases were separated by centrifugation (swinging rotor) at 1,500 x g for 5 min.
- The upper plasma membrane phase was carefully removed to a new tube, and mixed and repartitioned with equal weight of fresh lower phase solution (repeat one more time). Likewise, the lower phase (endosomal membrane phase) was repartitioned twice with equal weight of the upper phase.
- The final upper and lower phase were diluted, 3 fold and 10 fold, respectively, with 10 mM BTP-MES, pH 7.8, 1 mM EGTA, 1 mM EDTA, 0.29 M sucrose, and 20 mg/ml complete protease inhibitor cocktail, then centrifuged at 120,000 x g for 1 h.
- Pellets were resuspended in 10 mM BTP-MES, pH 7.8, 1 mM EGTA, 1 mM EDTA, 0.29 M sucrose, 20 mg/ml complete protease inhibitor cocktail.
- Microsomal membranes are prepared as section A above.
Recipes
- pH to 8.5
Grinding buffer Grinding buffer 100 ml 300 ml 0.29 M Sucrose 10 g 30 g 25 mM HEPES, pH =8.5 0.6 g 1.8 g 20mM EDTA (disodium salt) 0.74 g 2.23 g PVP(40,000) 0.5 g 1 g 0.2% BSA 0.2 g
Refrigerate
Just before use, add:3 mM DTT (1 M Stock) 300 μl 900 μl 200 ng/ml Leupeptin (200 μg/ml stock) 100 μl 300 μl PMSF (not AP assay) (100 mM in EtOH) 100 μl 300 μl 200 μM Benzamidine 100 μl 100 μl (from 200 mM each stock in EtOH) Pepstatin A (2 mg/ml STOCK) 100 μl Aprotinin (1 mg/ml STOCK) 10 μl 30 μl - --No GLYCEROL for membrane used in lipid raft purification
Resuspension buffer Resuspension buffer 100 ml 10 mM BTP-MES, pH 7.8 (1 M stock) 1 ml 250 mM sucrose 8.56 g 20% glycerol 20 g
Just before use, add:200 ng/ml Leupeptin (200 μg/ml stock) 100 μl PMSF (not AP assay) (1,000 mM in EtOH) 100 μl 200 μM benzamide/benzamidine (from 200 mM each stock in EtOH) 100 μl Pepstatin A (2 mg/ml STOCK) 100 μl Aprotinin (1 mg/ml STOCK) 10 μl - Stock solutions
20% (w/w) dextran in water
40% (w/w) polyethylene glycol 3350 in water
Incubate at 4 °C for 2-3 days - Phase mixture (2.7 g)
1.116 g 20% dextran
0.558 g 40% polyethylene glycol 3350
0.305 g sucrose
67.5 μl 0.2 M potassium phosphate (pH 7.8)
4.1 μl 2 M KCl
Add water to a final weight of 2.7 g - Phase system (30 g)
9.3 g 20% dextran
4.65 g 40% polyethylene glycol 3350
3.389 g sucrose
0.75 ml 0.2 M potassium phosphate (pH 7.8)
45 μl 2 M KCl
Add water to a final weight of 30 g
The phase system is mixed and allowed to settle overnight at 4 °C
The upper phase solution and the lower phase solution (for repartition) are collected
Stored separately at 4 °C
Acknowledgments
The work was supported by the Department of Energy, Basic Energy Sciences, grant no. DE-FG02-06ER15804 to ASM. HY was supported as part of the Center for Direct Catalytic Conversion of Biomass to Biofuels (C3Bio), an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, Award Number DE-SC0000997.
References
- Alexandersson, E., Gustavsson, N., Bernfur, K., Karlsson, A., Kjellbom, P. and Larsson, C. (2008). Purification and proteomic analysis of plant plasma membranes. Methods Mol Biol 432: 161-173.
- Blakeslee, J. J., Bandyopadhyay, A., Lee, O. R., Mravec, J., Titapiwatanakun, B., Sauer, M., Makam, S. N., Cheng, Y., Bouchard, R., Adamec, J., Geisler, M., Nagashima, A., Sakai, T., Martinoia, E., Friml, J., Peer, W. A. and Murphy, A. S. (2007). Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19(1): 131-147.
- Briskin, D. P. and Leonard, R. T. (1980). Isolation of tonoplast vesicles from tobacco protoplasts. Plant Physiol 66(4): 684-687.
- Hodges, T. K., Leonard, R. T., Bracker, C. E. and Keenan, T. W. (1972). Purification of an ion-stimulated adenosine triphosphatase from plant roots: association with plasma membranes. Proc Natl Acad Sci U S A 69(11): 3307-3311.
- Larsson, C., Widell, S. and Kjellbom, P. (1987). Preparation of high-purity plasma membranes. Methods Enzymol 148: 558-568.
- Leonard, R. T. and Vanderwoude, W. J. (1976). Isolation of plasma membranes from corn roots by sucrose density gradient centrifugation: an anomalous effect of ficoll. Plant Physiol 57(1): 105-114.
- Titapiwatanakun, B., Blakeslee, J. J., Bandyopadhyay, A., Yang, H., Mravec, J., Sauer, M., Cheng, Y., Adamec, J., Nagashima, A., Geisler, M., Sakai, T., Friml, J., Peer, W. A. and Murphy, A. S. (2009). ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J 57(1): 27-44.
- Yang, H., Richter, G. L., Wang, X., Mlodzinska, E., Carraro, N., Ma, G., Jenness, M., Chao, D. Y., Peer, W. A. and Murphy, A. S. (2013). Sterols and sphingolipids differentially function in trafficking of the Arabidopsis ABCB19 auxin transporter. Plant J 74(1): 37-47.
Materials and Reagents
Note: All chemicals were purchased from Sigma-Aldrich (http://www.sigmaaldrich.com/united-states.html) unless otherwise specified.
- Arabidopsis seedlings or mature tissue
- Ice
- Sucrose (Molecular Biology, Sigma-Aldrich, catalog number: 84097 )
- HEPES (Sigma-Aldrich, catalog number: H-3375 )
- EDTA (Research Products International, catalog number: E57020 )
- PVP (40,000) (Fisher Scientific, catalog number: BP431 )
- BSA (Sigma-Aldrich, catalog number: A-7906 )
- DTT (Sigma-Aldrich, catalog number: D0632 )
- Leupeptin (Sigma-Aldrich, catalog number: L2884 )
- PMSF (Sigma-Aldrich, catalog number: P-7626 )
- Benzamidine (Sigma-Aldrich, catalog number: B-6506 )
- Pepstatin A (Sigma-Aldrich, catalog number: P5318 )
- Aprotinin (Sigma-Aldrich, catalog number: A1153 )
- BTP-MES (BIS-TRIS propane, Sigma-Aldrich, catalog number: B6755 ; MES, Research Products International, catalog number: M22040 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- Dextran (GE, catalog number: 17-0320-01 )
- Polyethylene glycol 3350 (Union Carbide Corporation, catalog number: Carbowax 3350 )
- EGTA (Sigma-Aldrich, catalog number: E3889 )
- Protease inhibitor cocktail (Sigma-Aldrich, catalog number: P8340 )
- Grinding buffer (see Recipes)
- Resuspension buffer (see Recipes)
- Stock solutions (see Recipes)
- Phase mixture (see Recipes)
- Phase system (see Recipes)
Equipment
- Hermle Labnet Z383 centrifuge (Labnet International)
- OptimaTM-L-90K ultracentrifuge (Beckman Coulter)
- TLX centrifuge ( TLA 100.3 rotor) (Beckman Coulter)
- Waring blender jar (Waring Pro model: PBB212 )
- Mortar and pestle
- SW-28 rotor
- Nylon SS-34 tube
- SW-38 tube
- 3.5 ml polycarbonate TLA 100.3 tube
- 2 ml screwcap tubes
- 10, 15 and 50 ml Falcon tubes
- Polyallomer ultracentrifuge tube (Beckman Coulter)
Procedure
- Membrane preparation
- Turn on Hermle Labnet Z383 centrifuge. Set temperature to 4 °C. Collect 3-5 g of tissue from 5-d seedlings or mature tissue. Squeeze balls of tissue dry and weigh.
- For large quantity samples (> 40 g), add tissue to Waring blender jar and fill jar so that no air is present. Fill a bucket with ice. Pulse the waring jar for 30 s, then incubate in ice for 5 min. Repeat 3x.
- For small quantity samples (< 10 g), this step can be skipped, and the seedlings can be ground in grinding buffer in a mortar and pestle (see step A2).
- For large quantity samples (> 40 g), add tissue to Waring blender jar and fill jar so that no air is present. Fill a bucket with ice. Pulse the waring jar for 30 s, then incubate in ice for 5 min. Repeat 3x.
- Pour into pre-chilled motar and pestle on ice. Add a little bit of sand (acid washed, Sigma 18649).
- Immediately grind in a mortar and pestle for approximately 10 min.
- Pour through Miracloth lining a plastic power funnel set into nylon SS-34 tube. Squeeze through Miracloth to extract all of liquid homogenate. Fill tubes to 1 cm from top when standing upright.
- If the yield is poor, or the sample still look “unground”, it can be returned to mortar and pestle, and ground again in a small aliquot of grinding buffer.
- Immediately grind in a mortar and pestle for approximately 10 min.
- Balance the tubes and centrifuge at 8,000 x g, 4 °C for 15 min.
- Turn on Beckman Coulter OptimaTM-L-90K ultracentrifuge and set temperature to 4 °C.
- Carefully pour off supernatant from SS-34 tubes immediately without disturbing the pellet. Try not to collect any of the dark green pellet material in the supernatant fraction (the pellet is very easy to disturb).
- Collect the cellular debris pellet and store in liquid nitrogen until use, if desired.
- Turn on Beckman Coulter OptimaTM-L-90K ultracentrifuge and set temperature to 4 °C.
- To obtain a microsomal pellet, centrifuge the supernatant from the 8,000 x g spin at 100,000 x g, 4 °C, for 50 min, 1 h. Store additional supernatant on ice.
- Use thin wall tubes in the SW-28 rotor. The entire bucket assembly must be balanced within 0.005 g. Be sure to place buckets in the correct position and verify that they have engaged the rotor properly.
- Pour off supernatant and store at -70 °C (if soluble proteins are needed).
- If you have additional supernatant (from the 8,000 x g spin) on ice, pour off enough supernatant (from the 100,000 x g spin) from the ultracentrifuge tubes to allow for refilling with extra 8,000 x g supernatant. Balance tubes with poured off 100,000 x g supernatant as needed. Repeat 50 min spin.
- Use thin wall tubes in the SW-28 rotor. The entire bucket assembly must be balanced within 0.005 g. Be sure to place buckets in the correct position and verify that they have engaged the rotor properly.
- Pour off all supernatants–save as above if desired. Wipe out inside of tubes above the pellet with a Kimwipe or paper towel. Place on ice.
- Re-suspend each SW-28 pellet in microsomal resuspension buffer (volume will depend on pellet size and intended use, usually 1-2 ml). Use a cut-off 1 ml pipet tip to resuspend initially. This will be difficult, as the microsomes are very stick and hydrophobic. Try not to let the pellet stick to the pipet tip or walls of the centrifuge tube.
- After the pellet has been fairly well resuspended (pipet up and down for 100 times), use a normal 1 ml pipet tip to “break up” the clumps a bit and resuspend the microsomes more thoroughly (pipet up and down for 100 times).
- Finally, use a 200 μl pipet tip to break up the clumps and completely homogenize the preparation (pipet up and down for 200 times).
- If a wash of the pellet is desired, transfer the resuspended pellet to 3.5 ml polycarbonate TLA 100.3 tube. Wash each SW-38 tube with a bit of additional buffer, and add this to the TLA tubes. Centrifuge at 100,000 x g, 4 °C for 30 min in the TLX centrifuge (TLA 100.3 rotor). Discard supernatant, wipe out walls of tubes, store on ice.
- After the pellet has been fairly well resuspended (pipet up and down for 100 times), use a normal 1 ml pipet tip to “break up” the clumps a bit and resuspend the microsomes more thoroughly (pipet up and down for 100 times).
- Resuspend the microsomal pellet in resuspension buffer as in step A6. If fraction is being stored, aliquot into 500 μl–1 ml traction in 2 ml screwcap tubes. Freeze in liquid nitrogen, and store in either liquid nitrogen or -80 °C freezer until use.
- Turn on Hermle Labnet Z383 centrifuge. Set temperature to 4 °C. Collect 3-5 g of tissue from 5-d seedlings or mature tissue. Squeeze balls of tissue dry and weigh.
- Sucrose density gradients
Note: Preparation of sucrose solution: Important! Sucrose must be ultra pure.- Prepare 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55% (w/v) sucrose solution with 20 mM HEPES pH= 7.8, 1 mM EDTA in 10 ml tubes.
Note: Turn the tubes on a rotator for approximately 20 min until all sucrose has dissolved. - On the bottom of tube (Beckman polyallomer ultracentrifuge tube) put first the drop of 55% Suc and after that 2 ml of 50%, 45%, 40%, 35%, 30%, 25% and 20% were carefully added on the top, the interface should be seen (see Figure 1). At the top of the tube upon which to pipette the 0.5 ml sample of resuspended microsomal fraction (from membrane preparation).
Note: Allow sucrose to run down very slowly inside of tube. Sucrose layers should be visible when they are layering progressively. Place the tube with sucrose layers at 4 °C overnight to form continuous gradients.
Figure 1. A carton to show the interface of sucrose layers. All interfaces should be clearly seen between different concentrations. Please carefully add each concentration to obtain clear interface. - Centrifuge (Optima L-90K ultracentrifuge) 100,000 x g for 12 h.
- Immediately after the run the tube should be removed from the rotor taking care not to disturb the layers of sucrose gradient.
- Collect the 0.5 ml fractions from top to bottom very carefully to Eppendorf’s tubes.
- Prepare 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55% (w/v) sucrose solution with 20 mM HEPES pH= 7.8, 1 mM EDTA in 10 ml tubes.
- Two phase separation for plasma membrane and endomembrane fractions
Stock solutions, Phase mixture (2.7 g) and Phase system (30 g) (see Recipes)- Microsomal membranes are prepared as section A above.
- Fresh microsomal membranes are resuspended in 1 ml of buffer (0.33 M sucrose, 3 mM KCl, and 5 mM potassium phosphate, pH 7.8, and 20 mg/ml complete protease inhibitor cocktail from Sigma-Aldrich).
- Add 0.9 g resuspended membrane to 2.7 g two-phase separation mixture (can be scaled up).
- After mixing (inverting 30 times), the phases were separated by centrifugation (swinging rotor) at 1,500 x g for 5 min.
- The upper plasma membrane phase was carefully removed to a new tube, and mixed and repartitioned with equal weight of fresh lower phase solution (repeat one more time). Likewise, the lower phase (endosomal membrane phase) was repartitioned twice with equal weight of the upper phase.
- The final upper and lower phase were diluted, 3 fold and 10 fold, respectively, with 10 mM BTP-MES, pH 7.8, 1 mM EGTA, 1 mM EDTA, 0.29 M sucrose, and 20 mg/ml complete protease inhibitor cocktail, then centrifuged at 120,000 x g for 1 h.
- Pellets were resuspended in 10 mM BTP-MES, pH 7.8, 1 mM EGTA, 1 mM EDTA, 0.29 M sucrose, 20 mg/ml complete protease inhibitor cocktail.
- Microsomal membranes are prepared as section A above.
Recipes
- pH to 8.5
Grinding buffer Grinding buffer 100 ml 300 ml 0.29 M Sucrose 10 g 30 g 25 mM HEPES, pH =8.5 0.6 g 1.8 g 20mM EDTA (disodium salt) 0.74 g 2.23 g PVP(40,000) 0.5 g 1 g 0.2% BSA 0.2 g
Refrigerate
Just before use, add:3 mM DTT (1 M Stock) 300 μl 900 μl 200 ng/ml Leupeptin (200 μg/ml stock) 100 μl 300 μl PMSF (not AP assay) (100 mM in EtOH) 100 μl 300 μl 200 μM Benzamidine 100 μl 100 μl (from 200 mM each stock in EtOH) Pepstatin A (2 mg/ml STOCK) 100 μl Aprotinin (1 mg/ml STOCK) 10 μl 30 μl - --No GLYCEROL for membrane used in lipid raft purification
Resuspension buffer Resuspension buffer 100 ml 10 mM BTP-MES, pH 7.8 (1 M stock) 1 ml 250 mM sucrose 8.56 g 20% glycerol 20 g
Just before use, add:200 ng/ml Leupeptin (200 μg/ml stock) 100 μl PMSF (not AP assay) (1,000 mM in EtOH) 100 μl 200 μM benzamide/benzamidine (from 200 mM each stock in EtOH) 100 μl Pepstatin A (2 mg/ml STOCK) 100 μl Aprotinin (1 mg/ml STOCK) 10 μl - Stock solutions
20% (w/w) dextran in water
40% (w/w) polyethylene glycol 3350 in water
Incubate at 4 °C for 2-3 days - Phase mixture (2.7 g)
1.116 g 20% dextran
0.558 g 40% polyethylene glycol 3350
0.305 g sucrose
67.5 μl 0.2 M potassium phosphate (pH 7.8)
4.1 μl 2 M KCl
Add water to a final weight of 2.7 g - Phase system (30 g)
9.3 g 20% dextran
4.65 g 40% polyethylene glycol 3350
3.389 g sucrose
0.75 ml 0.2 M potassium phosphate (pH 7.8)
45 μl 2 M KCl
Add water to a final weight of 30 g
The phase system is mixed and allowed to settle overnight at 4 °C
The upper phase solution and the lower phase solution (for repartition) are collected
Stored separately at 4 °C
Acknowledgments
The work was supported by the Department of Energy, Basic Energy Sciences, grant no. DE-FG02-06ER15804 to ASM. HY was supported as part of the Center for Direct Catalytic Conversion of Biomass to Biofuels (C3Bio), an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, Award Number DE-SC0000997.
References
- Alexandersson, E., Gustavsson, N., Bernfur, K., Karlsson, A., Kjellbom, P. and Larsson, C. (2008). Purification and proteomic analysis of plant plasma membranes. Methods Mol Biol 432: 161-173.
- Blakeslee, J. J., Bandyopadhyay, A., Lee, O. R., Mravec, J., Titapiwatanakun, B., Sauer, M., Makam, S. N., Cheng, Y., Bouchard, R., Adamec, J., Geisler, M., Nagashima, A., Sakai, T., Martinoia, E., Friml, J., Peer, W. A. and Murphy, A. S. (2007). Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19(1): 131-147.
- Briskin, D. P. and Leonard, R. T. (1980). Isolation of tonoplast vesicles from tobacco protoplasts. Plant Physiol 66(4): 684-687.
- Hodges, T. K., Leonard, R. T., Bracker, C. E. and Keenan, T. W. (1972). Purification of an ion-stimulated adenosine triphosphatase from plant roots: association with plasma membranes. Proc Natl Acad Sci U S A 69(11): 3307-3311.
- Larsson, C., Widell, S. and Kjellbom, P. (1987). Preparation of high-purity plasma membranes. Methods Enzymol 148: 558-568.
- Leonard, R. T. and Vanderwoude, W. J. (1976). Isolation of plasma membranes from corn roots by sucrose density gradient centrifugation: an anomalous effect of ficoll. Plant Physiol 57(1): 105-114.
- Titapiwatanakun, B., Blakeslee, J. J., Bandyopadhyay, A., Yang, H., Mravec, J., Sauer, M., Cheng, Y., Adamec, J., Nagashima, A., Geisler, M., Sakai, T., Friml, J., Peer, W. A. and Murphy, A. S. (2009). ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J 57(1): 27-44.
- Yang, H., Richter, G. L., Wang, X., Mlodzinska, E., Carraro, N., Ma, G., Jenness, M., Chao, D. Y., Peer, W. A. and Murphy, A. S. (2013). Sterols and sphingolipids differentially function in trafficking of the Arabidopsis ABCB19 auxin transporter. Plant J 74(1): 37-47.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yang, H. and Murphy, A. (2013). Membrane Preparation, Sucrose Density Gradients and Two-phase Separation Fractionation from Five-day-old Arabidopsis seedlings. Bio-protocol 3(24): e1014. DOI: 10.21769/BioProtoc.1014.
Category
Plant Science > Plant biochemistry > Protein > Isolation and purification
Cell Biology > Organelle isolation > Membrane
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









