- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Zymogram Assay for the Detection of Peptidoglycan Hydrolases in Streptococcus mutans
Published: Vol 3, Iss 16, Aug 20, 2013 DOI: 10.21769/BioProtoc.855 Views: 15202
Reviewed by: Fanglian HeLin Fang

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

H2 Production from Methyl Viologen–Dependent Hydrogenase Activity Monitored by Gas Chromatography
Nuttavut Kosem
Dec 5, 2023 1770 Views

Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
John S. Smetana [...] Christopher W. Lennon
Apr 20, 2025 1585 Views

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1929 Views
Abstract
Peptidoglycan hydrolases or autolysins are enzymes capable of cleaving covalent bonds in bacterial peptidoglycan cell wall layer. They can participate in the cell division process, in the release of turnover products from peptidoglycan during cell growth, and in cell autolysis induced under particular conditions. The protocol for zymogram presented below should enable the identification of such enzymes through their separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis containing bacterial cells as substrate.
Keywords: PeptidoglycanMaterials and Reagents
- Bacterial strain (S. mutans UA159 wild-type strain or other S. mutans strains)
- Todd-Hewitt broth (BD Biosciences)
- Yeast-Extract (BioShop)
- Tris Base
- NaCl
- Sodium dodecyl sulfate (SDS)
- Triton X-100
- KOH
- MgCl2
- Glycine
- Glycerol
- Bromophenol blue
- Methylene blue
- 40% Acrylamide/Bis solution (37.5:1 acrylamide:bisacrylamide) (BioShop)
- Ammonium persulfate (Sigma-Aldrich)
- TEMED (BioBasic, Inc.)
- Ethanol
- Isopropanol
- dH2O
- Filter paper
- Precision Plus Protein Prestained Standards (Bio-Rad Laboratories)
- THYE broth (see Recipes)
- 20 mM Tris, 100 mM NaCl (pH 7.4) (see Recipes)
- 1.5 M Tris (pH 8.8) (see Recipes)
- 0.5 M Tris (pH 6.8) (see Recipes)
- SDS-PAGE loading buffer (see Recipes)
- Tris-Glycine SDS running buffer (see Recipes)
- Zymogram renaturing buffer (see Recipes)
- Staining solution (see Recipes)
- 10% separating gel solution (see Recipes)
- 4% stacking gel solution (see Recipes)
Equipment
- 15-ml canonical tubes
- Flasks
- 1.5 ml microcentrifuge tubes
- Refrigerated centrifuge
- Refrigerated microcentrifuge
- CO2 incubator
- Spectrophotometer
- Disposable plastic cuvettes
- Protein mini gel cassettes
- Heating block module
- Power supply
- Orbital shaker
- 37 °C temperature chamber
Procedure
- Preparation of the bacterial substrates incorporated into the gel
- Start 5 ml culture of S. mutans UA159 wild-type strain (or other S. mutans strains) in THYE broth into a 15-ml canonical tube and incubate overnight statically at 37 °C in air with 5% CO2.
- Inoculate 300 ml of fresh THYE broth with 1% of the overnight preculture into a 500-ml flask.
- Incubate the culture statically at 37 °C in air with 5% CO2 until an optical density at 600 nm (OD600) of 0.2 is reached.
- Harvest the cells by centrifugation at 10,000 x g for 10 min at 4 °C.
- Wash the cells using 5 ml of 20 mM Tris, 100 mM NaCl buffer (pH 7.4) and resuspend the cell pellet in 1.0 ml of 1.5 M Tris buffer (pH 8.8).
- Keep the cells at -20 °C until used.
- Start 5 ml culture of S. mutans UA159 wild-type strain (or other S. mutans strains) in THYE broth into a 15-ml canonical tube and incubate overnight statically at 37 °C in air with 5% CO2.
- Preparation of bacterial whole-cell extracts
- Start 5 ml overnight culture of S. mutans UA159 wild-type strain (or other S. mutans strains) in THYE broth into a 15-ml canonical tube and incubate statically at 37 °C in air with 5% CO2. Whole cell extract of a mutant strain deficient in the peptidoglycan hydrolase under study can also be analyzed concomitantly as negative control to confirm the specificity of the hydrolytic band(s) observed.
- Inoculate 10 ml of fresh THYE broth with 1% of the overnight preculture into a 15-ml canonical tube.
- Incubate the culture statically at 37 °C in air with 5% CO2 until the desired OD600 is reached. If the expression profile of the targeted peptidoglycan hydrolase is not known, we recommend to harvest cells at different optical densities corresponding to early log, mid-log, early stationary, and late stationary phase of growth.
- Harvest the cells by centrifugation at 10,000 x g for 10 min at 4 °C.
- Keep the cell pellet at -20 °C until used.
- Resuspend the cell pellet in 20 μl of SDS-PAGE loading buffer freshly prepared.
- Heat the samples at 95 °C for 10 min. Keep the samples on ice until loading.
- Start 5 ml overnight culture of S. mutans UA159 wild-type strain (or other S. mutans strains) in THYE broth into a 15-ml canonical tube and incubate statically at 37 °C in air with 5% CO2. Whole cell extract of a mutant strain deficient in the peptidoglycan hydrolase under study can also be analyzed concomitantly as negative control to confirm the specificity of the hydrolytic band(s) observed.
- Preparation of the zymogram gel
- Clean glass plates, spacers, and combs with ethanol and completely dry before use. Assemble the gel cassette following the manufacturer’s instructions.
- Prepare 10% separating gel solution (see Recipe 9).
- Transfer the separating gel solution (approx. 3.8 ml per small gel) to the casting chamber between the glass plates and fill up to about 0.7 cm below the bottom of the comb when the comb is in place.
- Add a small layer of isopropanol to the top of the gel prior to polymerization to straighten the level of the gel. Once the gel has polymerized, remove the isopropanol layer by several washes with dH2O, and dry with filter paper.
- Prepare 4% stacking gel solution (see Recipe 10).
- Pour the stacking gel solution (approx. 2.5 ml per small gel) on top of the separating gel until the space is full, and then insert the appropriate comb. Once the gel has polymerized, carefully remove comb.
- Remove the gel cassette from the casting stand and place it in the electrode assembly as recommended by the manufacturer.
- Pour Tris-Glycine SDS running buffer into the opening of the casting frame between the gel cassettes. Add enough buffer the fill the wells of the gel. Fill also the region outside of the frame.
- Load the samples (from Section II, step B-7) into each well as well as 5 μl of the Precision Plus Protein Prestained Standards.
- Connect the electrophoresis tank to the power supply. Run the gel at a constant voltage between 125-200 volts until the dye front is near the bottom of the gel.
- Clean glass plates, spacers, and combs with ethanol and completely dry before use. Assemble the gel cassette following the manufacturer’s instructions.
- Peptidoglycan hydrolase detection
- Remove the gel from the electrophoresis chamber, allow the gel to peel away and gently drop into a container. Wash the gel twice in 100 ml of dH2O for 30 min at room temperature under constant agitation.
- Incubate the gel in 100 ml of zymogram renaturing buffer for 30 min at room temperature under constant agitation. This step is necessary to renature the peptidoglycan hydrolases.
- Replace the zymogram renaturing buffer with fresh zymogram renaturing buffer and incubate the gel at 37 °C in a temperature chamber under constant agitation until clear hydrolytic band(s) appear, usually between 16 h and 48 h. The proteolytic activity appears as clear bands against a white background.
- Optional staining step: Decant the buffer and add 100 ml of staining solution, and incubate the gel at room temperature under constant agitation between 15 min and 2 h. Regions without staining are indicative of lysis (Figure 1). The proteolytic activity appears as clear bands against a blue background.
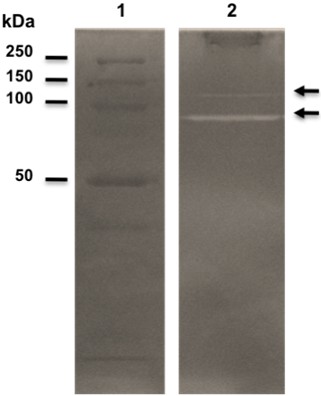
Figure 1. Zymogram activity gel after methylene blue staining. Heat-killed cells of S. mutans were used as substrate and were incorporated into a 10% SDS-PAGE gel. (1) Molecular size marker (Precision Plus Protein Prestained Standards). (2) Whole-cell extract of S. mutans UA159 wild-type strain. The two hydrolytic bands (arrows) observed correspond to the unprocessed (upper) and processed (lower) forms of the AtlA autolysin.
- Remove the gel from the electrophoresis chamber, allow the gel to peel away and gently drop into a container. Wash the gel twice in 100 ml of dH2O for 30 min at room temperature under constant agitation.
Recipes
- THYE broth
Dissolve 15 g of Todd-Hewitt and 1.5 g of Yeast Extract in 400 ml of dH2O
Once dissolved, bring up to a final volume of 500 ml with dH2O
Autoclave for 20 min at 120 °C
Store at room temperature
- 20 mM Tris, 100 mM NaCl buffer (pH 7.4)
Dissolve 2.42 g of Tris Base, and 5.84 g NaCl in 800 ml of dH2O
Once dissolved, adjust the pH to 7.4, and then bring up to a final volume of 1 L with dH2O
Store at 4 °C
- 1.5 M Tris pH 8.8 buffer
Dissolve 181.71 g of Tris Base in 800 ml of dH2O
Once dissolved, adjust the pH to 8.8, and then bring up to a final volume of 1 L with dH2O
Store at 4 °C
- 0.5 M Tris pH 6.8 buffer
Dissolve 60.57 g of Tris Base in 600 ml of dH2O
Once dissolved, adjust the pH to 6.8, and then bring up to a final volume of 1 L with dH2O
Store at 4 °C
- SDS-PAGE loading buffer (0.25 M Tris pH 6.8, 2% SDS, 10% glycerol, bromophenol blue)
Dissolve 0.3 g of Tris Base, 0.2 g SDS, 1.0 ml glycerol, and traces of bromophenol blue in 7 ml of dH2O
Once dissolved, bring up to a final volume of 10 ml with dH2O
- Tris-Glycine SDS running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS)
Dissolve 3.03 g of Tris Base, 14.4 g glycine, and 1 g SDS in 800 ml of dH2O
Once dissolved, bring up to a final volume of 1 L with dH2O
Store at 4 °C
- Zymogram renaturing buffer (20 mM Tris, 50 mM NaCl, 20 mM MgCl2, 0.5% Triton X-100, pH 7.4)
Dissolve 2.42 g of Tris Base, 2.92 g NaCl, and 4.06 g MgCl2 in 800 ml of dH2O
Adjust the pH to 7.4
Add 5 ml of Triton X-100, and bring up to a final volume of 1 L with dH2O
Store at 4 °C
- Staining solution (0.1% methylene blue, 0.01% KOH)
Dissolve 0.1 g of methylene blue and 0.01 g KOH in 100 ml of dH2O
Store at room temperature
- 10% separating gel solution
Mix the following reagents in a clean flask (total volume for 4 small gels):
7.4 ml dH2O
3.7 ml 40% acrylamide/bis
4 ml of bacterial substrate (from Section A, step A-6) boiled for 10 min just prior use
100 μl 10% SDS
50 μl 10% ammonium persulfate
5 μl TEMED
- 4% stacking gel solution
Mix the following reagents in a clean flask (total volume for 4 small gels):
6 ml dH2O
2.5 ml 0.5 M Tris (pH 6.8)
1.0 ml 40% acrylamide/bis
100 μl 10% SDS
100 μl 10% ammonium persulfate
25 μl TEMED
References
- Berg, K. H., Ohnstad, H. S. and Havarstein, L. S. (2012). LytF, a novel competence-regulated murein hydrolase in the genus Streptococcus. J Bacteriol 194(3): 627-635.
- Dufour, D. and Lévesque, C. M. (2013). Cell death of Streptococcus mutans induced by a quorum-sensing peptide occurs via a conserved streptococcal autolysin. J Bacteriol 195(1): 105-114.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Dufour, D. and Lévesque, C. M. (2013). Zymogram Assay for the Detection of Peptidoglycan Hydrolases in Streptococcus mutans. Bio-protocol 3(16): e855. DOI: 10.21769/BioProtoc.855.
Category
Microbiology > Microbial biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link