- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protein-peptide Interaction by Surface Plasmon Resonance
Published: Vol 3, Iss 3, Feb 5, 2013 DOI: 10.21769/BioProtoc.321 Views: 22715

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Analytical Gel Filtration for Probing Heavy Metal Transfer between Proteins
Steffen Lorenz Drees and Mathias Lübben
Aug 5, 2016 11109 Views

Hydrogen Deuterium Exchange Mass Spectrometry of Oxygen Sensitive Proteins
Luke Berry [...] Brian Bothner
Mar 20, 2018 10258 Views

Binding Affinity Quantifications of the Bacteriophage Mu DNA Modification Protein Mom Using Microscale Thermophoresis (MST)
Shubha Udupa [...] Shweta Karambelkar
Jul 20, 2022 3707 Views
Abstract
This protocol measures the protein-peptide interaction by surface plasmon resonance (SPR) using Biacore X100 (GE Healthcare). The Biacore system can monitor the direct interaction between biomolecules. There are several methods of immobilizing a ligand to the sensor chip. The optimal immobilization method for each experiment needs to be selected. In this protocol, we employed amine coupling to immobilize the protein to the carboxyl-type sensor chip. The procedure generally follows the “Instrument Handbook” of Biacore X100.
Keywords: SignalMaterials and Reagents
- Purified protein as ligand
- Synthetic peptide dissolved in DMSO as analyte
- NaCl
- NaOH
- HEPES
- Tween-20
- 50 mM NaOH
- Amine coupling kit {750 mg EDC (N-ethyl-N’-(3-dimethylaminopropyl)carbodiimide hydrochloride), 115 mg NHS (N-hydroxysuccinimide), 10.5 ml ethanolamine-HCl; GE Healthcare, catalog number: BR-10000-50 }
- 10 mM Acetate buffer pH 4 to 5.5 (GE Healthcare, catalog number: BR-1003-49 to 52 )
- DMSO (high grade, such as Sigma-Aldrich, catalog number: 276855 , or Sigma-Aldrich, catalog number: D-1435 )
- MilliQ Water
- Reagents for immobilization (see Recipes)
- Reagents for binding assay (see Recipes)
- Ligand (see Recipes)
- Analyte (see Recipes)
Equipment
- CM5 sensor chip (research grade; GE Healthcare, catalog number: BR-1003-99 )
- Biacore X100 Evaluation Software (GE Healthcare)
Procedure
- pH scouting (determination of the optimal pH for the ligand)
The positively charged ligand is electrostatically coupled to the negatively charged surface of the sensor chip, leading to ligand concentration. pH scouting is performed so as to determine the pH range that concentrates the ligand.
Dilute ligand to a final concentration of 5-200 μg/ml (higher protein concentration is necessary for ligands that are difficult to concentrate) in 10 mM acetate buffer of different pH. A regeneration step using 50 mM NaOH is performed after each pH scouting step.
Below is an example of pH scouting of a ligand to a sensor chip.
- Immobilization
Prepare the ligand at optimal pH and concentration. The optimal pH is the highest pH that allows ligand concentration in pH scouting. For example, pH5 is the optimal pH for the ligand shown in Figure 1.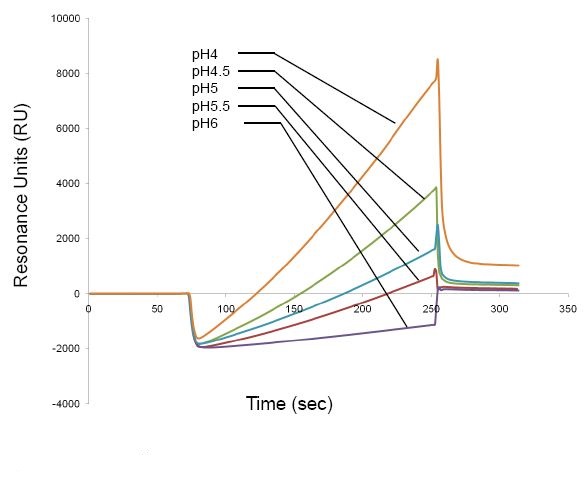
Figure 1. pH scouting
Below is an example of immobilization of a ligand to a sensor chip.
* EDC: N-ethyl-N’-(3-dimethylaminopropyl)carbodiimide hydrochloride
NHS: N-hydroxysuccinimide
These two and ethanolamine are provided in the Amine Coupling Kit
** Flow cell 1 serves as a reference cell.
After immobilization, run 1x running buffer for immobilization (1x RB-i) at 10 μl/min until the baseline becomes stable. A typical readout of immobilization is show in Figure 2.
Figure 2. Immobilization
- Analyte preparation
Precise sample preparation is required for accurate measurement, particularly when the analyte is dissolved in organic solvent. Even a subtle difference of organic solvent concentration between the sample and running buffer leads to inaccuracy.
Below is how we matched the DMSO concentration of our samples.- Dilute 10x HBS buffer with MilliQ water and Tween-20 to 1.1x HBS buffer with 0.11% Tween-20.
- Add 25 ml of DMSO and 25 ml of MilliQ water to 450 ml of 1.1x HBS buffer with 0.11% Tween-20 to make 1x running buffer for binding assay (1x RB-b).
- Dilute 10 mM peptide in DMSO to 5 mM with MilliQ water, and further dilute to 500 μM with 1.1x HBS buffer with 0.11% Tween20 (this makes 500 μM peptide in 1x RB-b).
- Dilute 500 μM peptide mixture to 12.5 μM with 1x RB-b, followed by two-fold dilution to 0.39 μM with 1x RB-b.
- Dilute 10x HBS buffer with MilliQ water and Tween-20 to 1.1x HBS buffer with 0.11% Tween-20.
- Regeneration condition
For analytes with slow dissociation (when the sensorgram does not go back to the background level at the end of the dissociation step), a regeneration step must be performed after each binding assay. Determine an optimum regeneration condition for each analyte. The regeneration should not change the activity of the immobilized ligand (that is, the responses obtained from the binding assays before and after regeneration should be the same). - Binding assay
Perform the binding assay was with 1x RB-b.
Below is an example of a binding assay.
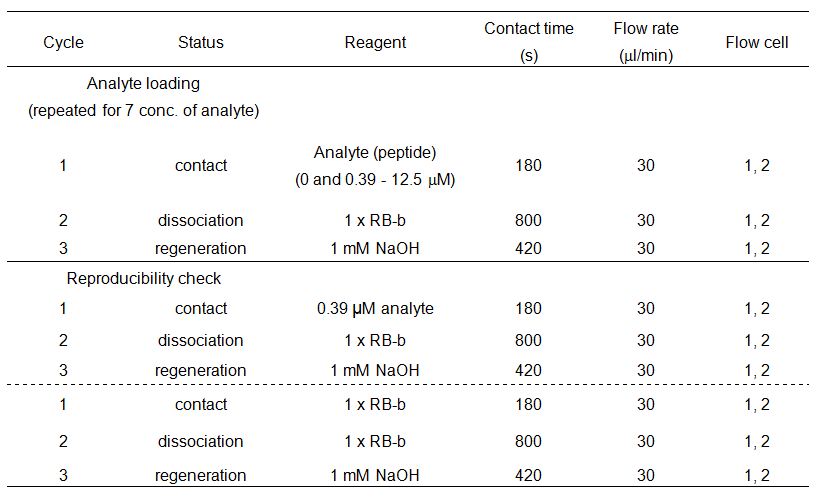
Data analysis is performed with the Biacore X100 Evaluation Software (GE Healthcare).
A sensorgram of protein-peptide interaction is shown in Figure 3 and also in the reference (Eguchi et al., 2012).
Figure 3. Binding assay
Recipes
The buffers used for immobilization and binding must be optimized for every ligand and analyte. The following recipe worked with our protein and peptide samples.
- Reagents for immobilization
1x running buffer for immobilization (1x RB-i)
10 mM HEPES (pH 7.5)
150 mM NaCl
0.05% Tween-20 - Reagents for binding assay
10x HBS buffer [100 mM HEPES (pH7.5), 1.5 M NaCl]
1x running buffer for binding assay (1x RB-b)
10 mM HEPES (pH7.5)
150 mM NaCl
0.1% Tween-20
5% DMSO - Ligand
Purified protein (purity > 90%) equilibrated in 1x RB-i. - Analyte
Synthetic peptide dissolved in DMSO.
Acknowledgments
This protocol was adapted from Eguchi et al. (2012), and generally follows the “Instrument Handbook” of Biacore X100 (GE Healthcare). This work was supported by a Grant-in-Aid for Scientific Research (A, 20248012) from the Japan Society for the Promotion of Science (JSPS), the Research and Development Program for New Bio-Industry Initiatives (2006–2010) of Bio-Oriented Technology Research Advancement Institution (BRAIN), Japan, MEXT-Supported Program for the Strategic Research Foundation at Private Universities, 2011-2015 (S1101035), Sasakawa Scientific Research Grant from The Japan Science Society, and the Institute for Fermentation, Osaka.
References
- Eguchi, Y., Ishii, E., Yamane, M. and Utsumi, R. (2012). The connector SafA interacts with the multi-sensing domain of PhoQ in Escherichia coli. Mol Microbiol 85(2): 299-313.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ishii, E., Eguchi, Y. and Utsumi, R. (2013). Protein-peptide Interaction by Surface Plasmon Resonance. Bio-protocol 3(3): e321. DOI: 10.21769/BioProtoc.321.
Category
Biochemistry > Protein > Interaction > Protein-ligand interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










