- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Rubisco Extraction and Purification from Diatoms
Published: Vol 7, Iss 6, Mar 20, 2017 DOI: 10.21769/BioProtoc.2191 Views: 11961
Reviewed by: Scott A M McAdamAgnieszka ZienkiewiczAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
![Assessing Long-distance Transport from Photosynthetic Source Leaves to Heterotrophic Sink Organs with [<sup>14</sup>C]CO<sub>2</sub>](https://en-cdn.bio-protocol.org/imageup/arcimg/20171211011113752.jpg?t=1773180253)
Assessing Long-distance Transport from Photosynthetic Source Leaves to Heterotrophic Sink Organs with [14C]CO2
Umesh P Yadav [...] Brian G Ayre
Dec 20, 2017 6602 Views
![Quantifying the Capacity of Phloem Loading in Leaf Disks with [<sup>14</sup>C]Sucrose](https://en-cdn.bio-protocol.org/imageup/arcimg/20171211011350163.jpg?t=1773180253)
Quantifying the Capacity of Phloem Loading in Leaf Disks with [14C]Sucrose
Umesh P Yadav [...] Brian G Ayre
Dec 20, 2017 8050 Views

13CO2-labelling and Sampling in Algae for Flux Analysis of Photosynthetic and Central Carbon Metabolism
Or Geffen [...] Haim Treves
Sep 5, 2023 1980 Views
Abstract
This protocol describes a method to extract ribulose-1,5-bisphosphate carboxylase oxygenase (Rubisco) from diatoms (Bacillariophyta) to determine catalytic performance. This protocol has been adapted from use in cyanobacteria and higher plants (Andrews, 1988; Whitney and Sharwood, 2007). First part (steps A1-A3) of the extraction provides a crude extract of Rubisco that is sufficient for carboxylation assays to measure the Michaelis constant for CO2 (KC) and the catalytic turnover rate (kcatc). However, the further purification steps outlined (steps B1-B4) are needed for measurements of Rubisco CO2/O2 Specificity (SC/O, [Kane et al., 1994]).
Keywords: RubiscoBackground
Ribulose-1,5-bisphosphate carboxylase oxygenase (Rubisco, EC 4.1.1.39) catalyzes the first step in the photosynthetic assimilation of CO2 and thus plays a fundamental role in photosynthesis and the global carbon cycle. Rubisco has been isolated from a wide range of organisms, from archaea, bacteria, algae to plants, and displays a diverse range of kinetics between organisms (Galmes et al., 2014; Tcherkez et al., 2006; Whitney et al., 2011). Knowledge of Rubisco kinetics is a key component for understanding how photosynthesis and thus the biological sink of carbon will respond to rising anthropogenic CO2. Diatoms are a group of unicellular algae responsible for ~20% of global photosynthesis (Falkowski and Raven, 2007) but as yet have been relatively poorly studied in terms of their Rubisco kinetics.
Isolation and purification of Rubisco is required before kinetic assays can be undertaken. Due to differences in cell structure and organic composition between organisms, the method for the purification of viable Rubisco enzyme needs to be continually optimized. This protocol describes a method to extract and purify Rubisco using size exclusion chromatography from diatoms in preparation for kinetic assays. The method is similar to Whitney and Sharwood (2007), used for the purification of Rubisco overexpressed in E. coli, in that a French press is used to mechanically rupture cells. The French press is necessary to obtain sufficient cell lysis as diatoms are unicellular with silica frustules, unlike plant tissue in which sufficient lysis is easily achieved by homogenizing frozen leaf tissue in a mortar and pestle. Furthermore, due to the low in vivo concentrations of Rubisco in diatoms (Losh et al., 2013), large diatom culture volumes concentrated via centrifugation, are needed to obtain enough biomass compared to plant tissue and E. coli lines with overexpressed Rubisco.
Materials and Reagents
- 15 ml centrifuge tubes with conical bottoms
- 50 ml centrifuge tubes with conical bottoms
- 500 ml centrifuge tubes with conical bottoms
- 1.5 ml microcentrifuge tubes
- 1 ml syringe
- 1 ml Bio-Scale mini Macro-Prep high Q ion exchange column (Bio-Rad Laboratories, catalog number: 7324120 )
- Amicon Ultra-4 centrifugal filter (30,000 NMWL) (EMD Millipore, catalog number: UFC803024 )
- Amicon Ultra-100 centrifugal filter (100,000 NWML) (EMD Millipore, catalog number: UFC910024 )
- Diatoms
- Liquid nitrogen
- Polyvinylpolypyrrolidone (PVPP; insoluble) (Sigma-Aldrich, catalog number: 77627 )
- Additional materials To test for Rubisco activity (Optional):
Labelled CO2 (as NaH14CO3) (5 mCi) (PerkinElmer, catalog number: NEC086H005MC )
Ribulose-1,5-bisphosphate (RuBP; synthesized, purified and stored anaerobically as described in Kane et al., 1998) - Acetic acid (Sigma-Aldrich, catalog number: A9967 or 27225 )
Note: The product acetic acid ( A9967 ) has been discontinued. - Methanol (Sigma-Aldrich, catalog number: M1770 or 494437 )
Note: The product Methanol ( M1770 ) has been discontinued. - Soluble protein (as determined by Bradford assay) (Coomassie Plus Assay Kit) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 23236 )
- Additional materials to test for protein using Native and SDS-PAGE (Optional):
Gel code blue stain (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 24590 )
4-12% Tris-glycine mini gels (Thermo Fisher Scientific, InvitrogenTM, catalog number: XV04120PK20 )
4-12% Bis-Tris gels (Thermo Fisher Scientific, InvitrogenTM, catalog number: NP0321PK2 )
SDS reducing buffer for SDS-PAGE (see Recipes)
TBS buffer for SDS-PAGE (see Recipes)
AttoPhos reagent (Astral Scientific, Gymea, NSW, Australia)
Antisera raised against the large subunit holoenzyme of Rubisco in Phaeodactylum tricornutum
Alkaline Phosphatase conjugated secondary antibody - 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid (EPPS) (Sigma-Aldrich, catalog number: E9502 )
- Ethylenediaminetetraacetic acid disodium salt (EDTA) (Sigma-Aldrich, catalog number: E5134 )
- Dithiothreitol (Sigma-Aldrich, catalog number: D0632 )
- Plant protease inhibitor cocktail (Sigma-Aldrich, catalog number: P9599 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653 )
- Triethanolamine (Sigma-Aldrich, catalog number: 90279 )
- Magnesium acetate (Sigma-Aldrich, catalog number: M5661 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- Extraction buffer (see Recipes)
- Column buffer (see Recipes)
- Column elution buffer (see Recipes)
- Specificity (SC/O) buffer (see Recipes)
Note: All chemicals are of A.C.S. grade.
Equipment
- French pressure cell press (Thermo Fisher Scientific, model: FA-078 )
- Coulter Counter Z Series (Beckman Coulter, model: Z Series Coulter Counter )
- Centrifuge (large volumes [1 L] at 2,000 x g, small volumes [< 15 ml] at 17, 600 x g, 4 °C)
- Fume hood
- Superdex 200 (GE Healthcare, catalog number: 17517501 )
- FPLC (Äkta Pure 25) setup at 4 °C for size-exclusion chromatography using Superdex 200/30 (GE Healthcare, model: Äkta Pure 25 )
- To confirm purification and activity of Rubisco:
- Protein transfer apparatus and immunoblot imagining equipment – to check abundance and purity of extracted Rubisco
- Radioisotope laboratory and associated septum capped vials and syringes
Procedure
- Total soluble cellular (crude) protein extraction – suitable for measurement of the Michaelis constant for CO2 (KC), catalytic turnover rate (kcatc) and the first step for CO2/O2 specificity assays (SC/O).
- Diatoms are grown in 2 L batch cultures at 20 °C under continuous light (c. 150 μmol photons m-2 sec-1) in sterile seawater to which 0.2 μM filtered nutrients, vitamins and trace metals were added to give the final concentrations as defined in the Aquil medium. For a full recipe and guidelines for making Aquil recipe, see Sunda et al., 2005. Growth rates are monitored by cell counting using a Coulter Counter. During exponential growth, cells are concentrated to a pellet via gentle centrifugation (2,000 x g for 10 min) at 15 °C and the supernatant discarded. Approximately 1 g of a cell pellet is required to purify Rubisco for measuring SC/O. To limit storage space, collect cells into 1.5 ml microcentrifuge tubes and then snap freeze in liquid nitrogen and store at -80 °C until further analysis.
- The cell pellets are re-suspended in a total of 5 ml ice cold extraction buffer in a 15 ml polypropylene tube. The cells are ruptured using a pre-chilled French press at 140 MPa (see Figure 1 for photo and Video 1 of French press) and the cell lysate collected into the same tube.

Figure 1. Cell lysis using a French press. Photo of a French Pressure Cell that can efficiently lyse microalgae cells, including diatoms. Algae cell extracts in ice cold buffer are placed in the ice-cold stainless steel French Pressure Cell and hydraulic pressure applied. Once at 140 MPa the outlet to the Cell is slowly opened. The cells lyse as they emerge from high to ambient pressure and the cell extract is collected into a 15 ml or 50 ml polypropylene tubes. A slow flow rate of sample out of the Cell (~1 to 2 drops sec-1) is maintained to ensure the pressure is maintained (see Video 1, acknowledgement Bratati Mukherjee).Video 1. Use of French press to lyse cells - To the cell lysate, add polyvinylpolypyrrolidone (1%, w/v) to bind secondary metabolites, which are then removed by centrifugation (17,600 x g, 4 °C, 5 min).
- After centrifugation Rubisco remains fully intact as confirmed by native PAGE (see Data analysis, Figure 2) and fully soluble, remaining in the cell lysis supernatant as confirmed by SDS-PAGE (see Data analysis, Figure 3).
- Rubisco kcatc (and KC) is quantified by 14CO2 fixation assays (see Data analysis).
- Rubisco in the soluble protein extract is activated for 10 to 15 min with 15 mM NaH14CO3 and 15 mM MgCl2.
- In 7 ml septum capped glass scintillation vials 20 μl of the extract is assayed for 60 sec in 0.5 ml of 50 mM EPPS-NaOH pH 8.2, 10 mM MgCl2, 0.6 mM RuBP, 10 μg ml-1 carbonic anhydrase and varying NaH14CO3 concentrations (1.2 to 12 mM which equates to ~15 to 150 μM 14CO2 at pH 8.0 at 25 °C) (Sharwood et al., 2016). The buffer and vials are equilibrated with the appropriate O2/N2 gas mixture prior to adding the NaH14CO3 and sample.
- The reaction is stopped with 0.2 ml 0.5 N formic acid and non-fixed 14CO2 is vented by heating the vials at 85 °C in the fume hood.
- The dried residue is dissolved in 0.5 ml double distilled H2O and vortexed with 1 ml Scintillant (Packard, Ultima Gold XR) before the labelled 3-phosphoglycerate (14C) is measured in a scintillation counter. This crude extraction is suitable for measurements of Rubisco maximum carboxylation rate (Vmax) and Rubisco Michaelis constant for CO2 under N2 (KC) and 21:79% O2:N2 (Kcair) (Sharwood et al., 2008; Sharwood et al., 2016). To quantify kcatc the Vmax value is divided by the Rubisco content in the assay. Measuring Rubisco content is achieved using the 14C-CABP binding method or by immunoblot analysis. The accuracy and experimental limitations of both approaches are detailed in Whitney and Sharwood, 2014. Purification of Rubisco is necessary for measuring SC/O (see next step).
- Diatoms are grown in 2 L batch cultures at 20 °C under continuous light (c. 150 μmol photons m-2 sec-1) in sterile seawater to which 0.2 μM filtered nutrients, vitamins and trace metals were added to give the final concentrations as defined in the Aquil medium. For a full recipe and guidelines for making Aquil recipe, see Sunda et al., 2005. Growth rates are monitored by cell counting using a Coulter Counter. During exponential growth, cells are concentrated to a pellet via gentle centrifugation (2,000 x g for 10 min) at 15 °C and the supernatant discarded. Approximately 1 g of a cell pellet is required to purify Rubisco for measuring SC/O. To limit storage space, collect cells into 1.5 ml microcentrifuge tubes and then snap freeze in liquid nitrogen and store at -80 °C until further analysis.
- Rubisco purification for measuring SC/O
The soluble cellular protein needs to be further purified before quantifying SC/O (CO2/O2 specificity).- Soluble cellular protein from step A3 above is manually passed by syringe through a 1 ml Bio-Scale mini Macro-Prep high Q ion exchange (IEX) column equilibrated with column buffer. All steps are performed at 4 °C. The method involves: Pre-washing the IEX column is with 2 ml of column buffer. Soluble cellular protein is passed through the column by syringe (flowrate at ~3 to 5 ml min-1) and then the column is washed with 8 ml of column buffer. Bound Rubisco is eluted in 1.5 ml column elution buffer, collecting in three 500 μl elution fractions. The last two fractions contain > 90% of the Rubisco activity and are therefore pooled before concentrating to ~0.4 ml by centrifugation (4,000 x g, 10 min, 4 °C) using an Amicon Ultra-100 centrifugal filter (i.e., a 100 kDa MW cut-off filter).
- The concentrated Rubisco is injected into the 200 μl sample loop of the Äkta purification system onto a Superdex 200 column pre-equilibrated with SC/O buffer at 4 °C. The sample is loaded onto the column using the flow rate of 0.5 ml/min (max delta column pressure alarm set to 1.5 MPA) and fractions (0.5 ml) collected after the void volume (8 ml). Absorbance at 280 nm and conductance are continuously monitored. RuBP dependent 14CO2-fixation activity in the fractions is assayed and those with peak Rubisco activity (typically fractions 5 to 7) are pooled and concentrated to ~0.1 ml by centrifugation (4,000 x g, 20 min, 4 °C) using an Amicon Ultra-100 centrifugal filter.
- The concentrated and purified Rubisco is now ready for the SC/O assay. Alternatively, glycerol can be added to 20% (v/v) final concentration and the enzyme then frozen in liquid nitrogen and stored at -80 °C.
- Optional: the SC/O assay (not part of this protocol on Rubisco extraction)
- SC/O assays are carried out at 25 °C (or other temperature) according to the method of (Kane et al., 1994). The purified Rubisco (10-50 μl) is injected into 20 ml glass, septum sealed glass vials containing 1 ml assay buffer (see Recipes) that has been equilibrated for 0.5 to 1 h with 500 ppm CO2 mixed with O2 using Wostoff gas-mixing pumps. After 15 min, further equilibration the reactions are initiated with the injection of 1 nmol of 2-3H-RuBP. Alkaline phosphatase is injected after 30 min to convert the 3-3H-phosphoglycerate (carboxylation product) and 2-3H-phosphoglycolate (oxygenation product) into 3H-3-glycerate and 3H-2-glycolate.
- The reactions are passed through a 0.4 ml AGI-X8 (10% formate) anion-exchange column and then the bound 3H-3-glycerate and 3H-2-glycolate eluted in 0.5 ml 20% (v/v) H2SO4 before separating them by HPLC using an isocratic gradient of 0.012 M H2SO4 through a Aminex HPX-87H column at 65 °C at a flow rate of 0.4 ml min-1.
- The SC/O at 25 °C is then calculated from the radioactivity in the 3H-3-glycerate and 3H-2-glycolate using equation:
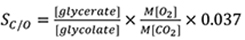
Where,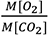 represents the molar ratio of O2 and CO2 (99.95% and 0.05% [v/v] respectively),
represents the molar ratio of O2 and CO2 (99.95% and 0.05% [v/v] respectively),
0.037 is the proportion of the CO2 solubility relative to O2 in H2O at 25 °C (Kane et al., 1994).
- SC/O assays are carried out at 25 °C (or other temperature) according to the method of (Kane et al., 1994). The purified Rubisco (10-50 μl) is injected into 20 ml glass, septum sealed glass vials containing 1 ml assay buffer (see Recipes) that has been equilibrated for 0.5 to 1 h with 500 ppm CO2 mixed with O2 using Wostoff gas-mixing pumps. After 15 min, further equilibration the reactions are initiated with the injection of 1 nmol of 2-3H-RuBP. Alkaline phosphatase is injected after 30 min to convert the 3-3H-phosphoglycerate (carboxylation product) and 2-3H-phosphoglycolate (oxygenation product) into 3H-3-glycerate and 3H-2-glycolate.
- Soluble cellular protein from step A3 above is manually passed by syringe through a 1 ml Bio-Scale mini Macro-Prep high Q ion exchange (IEX) column equilibrated with column buffer. All steps are performed at 4 °C. The method involves: Pre-washing the IEX column is with 2 ml of column buffer. Soluble cellular protein is passed through the column by syringe (flowrate at ~3 to 5 ml min-1) and then the column is washed with 8 ml of column buffer. Bound Rubisco is eluted in 1.5 ml column elution buffer, collecting in three 500 μl elution fractions. The last two fractions contain > 90% of the Rubisco activity and are therefore pooled before concentrating to ~0.4 ml by centrifugation (4,000 x g, 10 min, 4 °C) using an Amicon Ultra-100 centrifugal filter (i.e., a 100 kDa MW cut-off filter).
Data analysis
- Intact Rubisco remains in the soluble fraction during extraction
It was determined that full cell lysis is routinely obtained using a French Pressure cell and that Rubisco remains intact and within the soluble fraction during the crude extraction (steps A1-A3). Native PAGE (Figure 2) shows Rubisco remains as an intact L8S8 enzyme (comprising 8 large [L] and 8 small [S] subunits), even following electrophoresis for 16 h at 4 °C and 60 V. The differing mobility of the Rubsico band between species occurs due to subunit sequence differences and subtle variations in quaternary structural properties (i.e., slight differences in amino acid sequence and length). SDS-PAGE (Figure 3) demonstrates that comparable levels of Rubisco L- and S-subunit are detected in the whole cell lysis (after mechanical rupture but prior to centrifugation, step A2) and soluble cell protein (after centrifugation, step A3) indicating the extracted Rubisco is fully soluble. - Rubisco retains activity after extraction
The activity of Rubisco within the crude extracts (steps A1-A3) of 11 diatom species were tested and published (Young et al., 2016). Full activation of Rubisco was obtained after 10 min of extraction at 25 °C and the activity remained stable during a further 10 min testing. Measurements of KC and kcatc for the diatom Rubiscos are published in (Young et al., 2016). - Quantifying SC/O (steps B1-B4). Figure 4 shows the 3H-elution profile of the HPLC separated 3H-glycerate and 3H-glycolate peaks which are used to calculate SC/O as described above in step B4c.
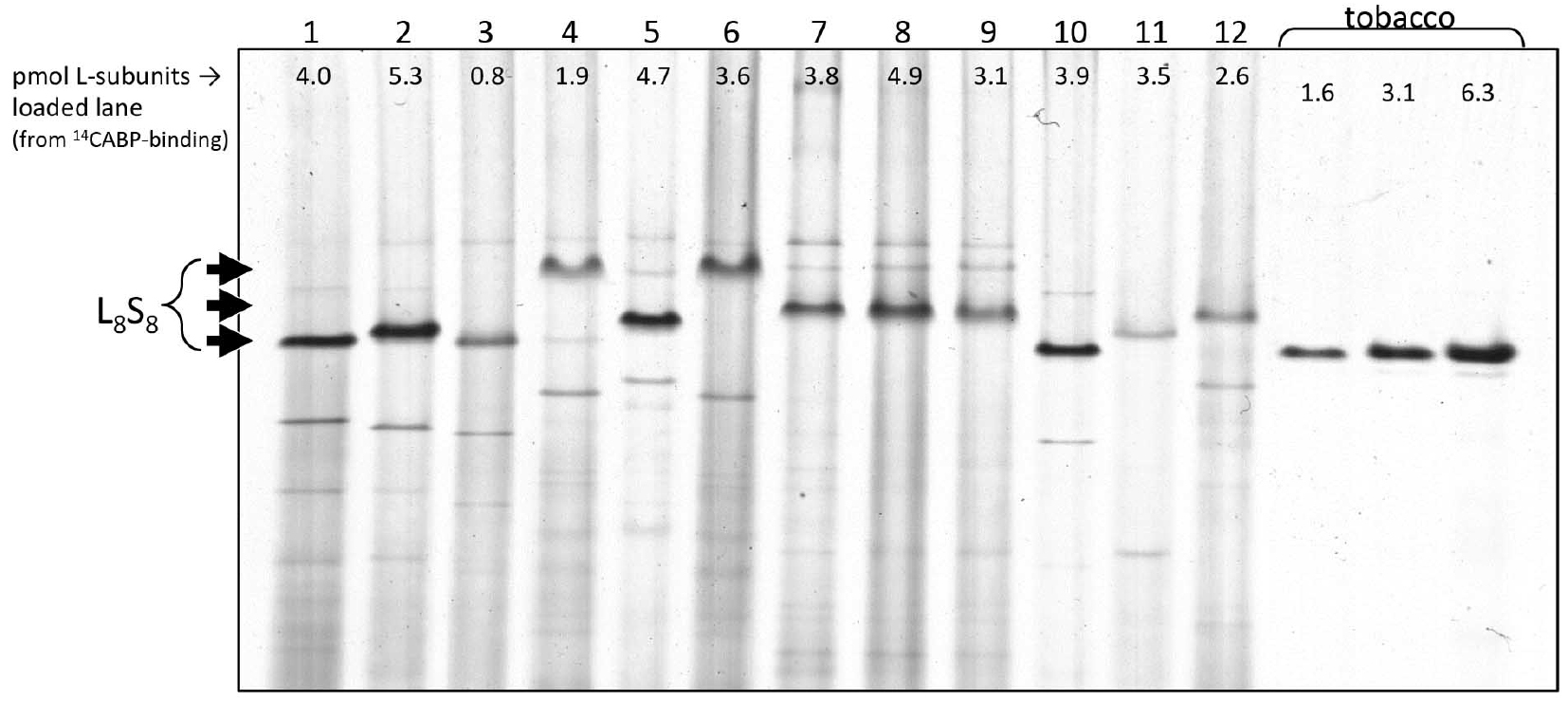
Figure 2. Native-PAGE blot of crude extract to show fully intact Rubisco protein. Total soluble cell extract (5 μg as determined by Bradford assay, Coomassie plus) was separated at 4 °C through a precast 4-12% Tris-glycine gel overnight (16 h) at 60 V. Gel was rinsed with deionized water, fixed for 30 min with 45% (v/v) H2O, 5% (v/v) acetic acid and 50% (v/v) methanol the extensively rinsed with multiple changes if deionized water. The proteins were visualized using Gelcode blue Coomassie stain (Invitrogen). Arrows indicate where the ~520 kDa Rubisco complex (L8S8) locates on the gel. Lane numbers indicate extract from different diatom species: (1) Thalassiosira weissflogii CCMP 1336, (2) Thalassiosira oceania CS-427, (3) Skeletonema marinoi CCMP 1332, (4) Chaetoceros calcitrans CCMP 1315, (5) Chaetoceros calcitrans CS-178, (6) Chaetoceros muelleri CCMP 1316, (7) Phaeodactylum tricornutum CCMP 642, (8) Phaeodactylum tricornutum UTEX 630, (9) Phaeodactylum tricornutum CS-29, (10) Bellerochea sp. CS-874/01, (11) Isochrysis sp. CS-177, (12) Pleurochrysis cartera CS-287. Last three lanes contain 5, 10 and 20 μl of crude cell extract from tobacco as a control. Shown are the Rubisco active site contents quantified for each sample by 14C-CABP binding (Whitney and Sharwood, 2014; Sharwood et al., 2008).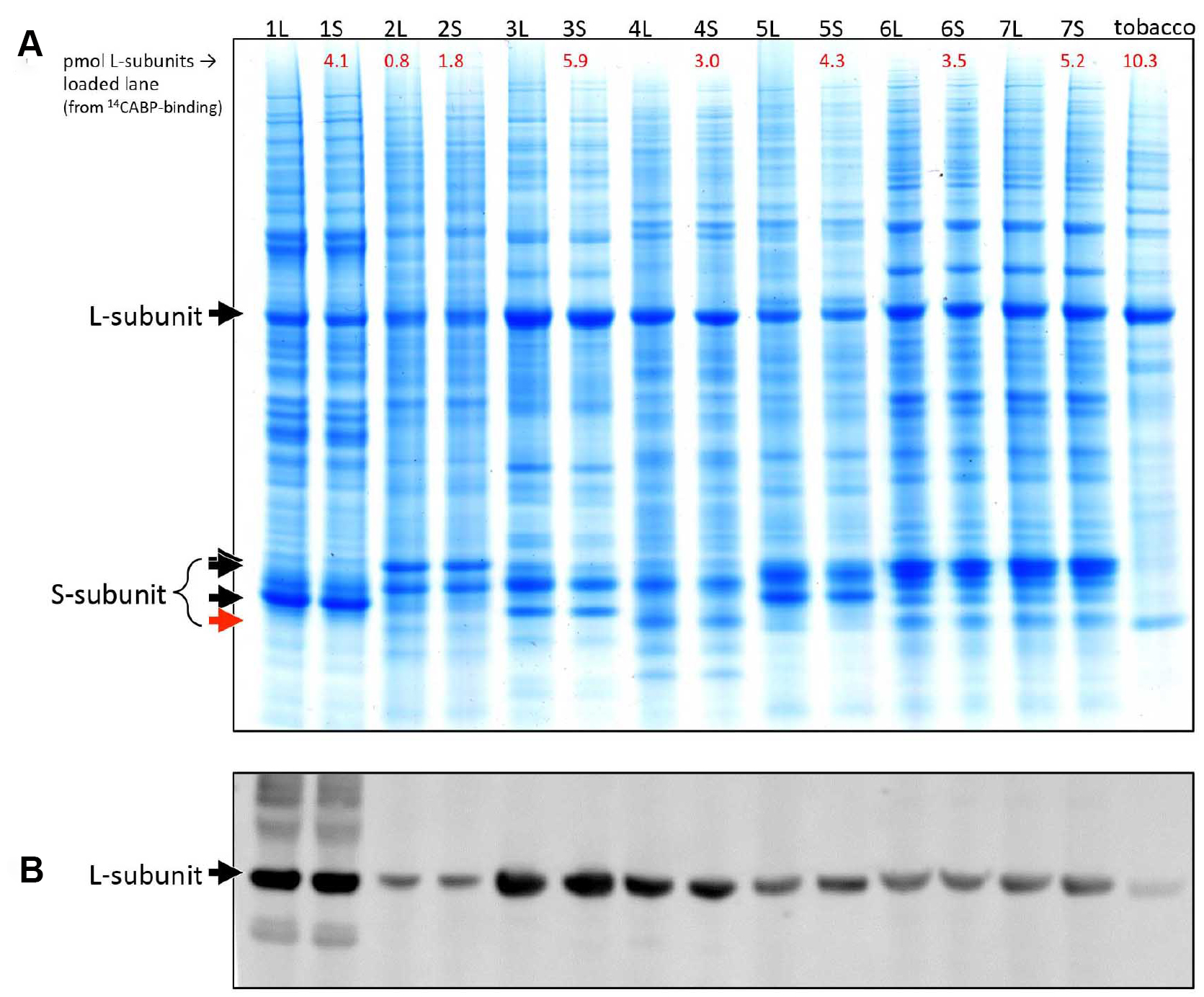
Figure 3. SDS-PAGE analysis of Rubisco solubility, integrity and complete extraction by French Pressure Cell lysis. A. Coomassie stain and B. Diatom Rubisco antibody blot (see Whitney et al., 2001 for details) of total cellular lysate (L, following French Pressure Cell lysis) and soluble cellular protein (S, following centrifugation) from the diatom species: (1) P. tricornutum CS-29, (2) Skeletonema ardens CS-348, (3) Pavlova lutheri CS-182, (4) Fragilariopsis cylindrus CCMP 1102, (5) Cylindrotheca fusiformis CS-13, (6) Thalassiosira oceania CS-427, and (7) Thalassiosira weissflogii CCMP 1336. 5 μl of tobacco soluble leaf protein was loaded for comparison. The equal intensity of the L-subunit in both the L and S protein fractions indicate all the Rubisco was extracted (complete cell lysis) and fully soluble with no L-subunit degradation evident in the Western blot. Sample preparation and electrophoresis: protein (L or S) extracts (150 μl) were added to 4x SDS-reducing buffer (50 μl) and boiled for 5 min then centrifuged (16,000 x g, 5 min) before separating by 4-12% Bis-Tris SDS-PAGE at 200 V for 45 min in MES buffer (50% methanol, 40% H2O and 10% glacial acetic acid). Duplicate gels were either (A) fixed and Coomassie stained (see Figure 2) or (B) the proteins transferred onto nitrocellulose membrane and probed with an antibody to P. tricornutum Rubisco (see Whitney et al., 2001 for further experimental details).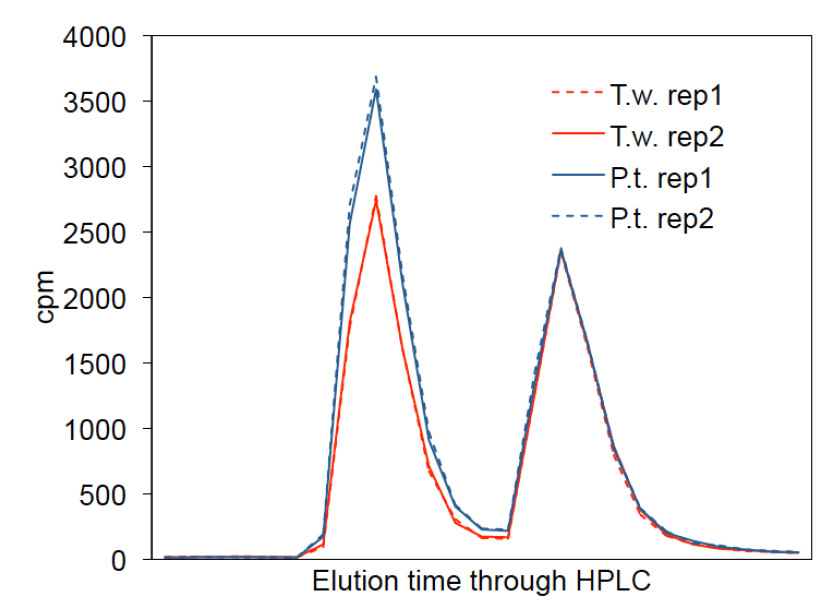
Figure 4. 3H-Elution profile of the HPLC fractions that are used to calculate SC/O from the amount of 3H incorporated into 3H-glycerate and 3H-glycolate (peaks 1 and 2 respectively) as described in step B4c. Shown here are results of an SC/O assay using purified Rubisco from the diatoms, Thalassiosira weissflogii (T.w., red) and Phaeodactylum tricornutum (P.t., blue) showing two technical replicates (rep) for each.
Recipes
- Extraction buffer
50 mM EPPS-NaOH, pH 8.0
1 mM EDTA
2 mM dithiothreitol (DTT)
1% (v/v) plant protease inhibitor cocktail
Dissolve EPPS and EDTA in deionized water to the final desired concentrations
Bring pH to 8.0 with NaOH
Notes:- Add DTT just prior to starting extractions.
- Immediately prior to use, add protease inhibitor cocktail to the aliquot of buffer being used for extraction.
- Add DTT just prior to starting extractions.
- Column buffer
50 mM EPPS-NaOH, pH 8.0
1 mM EDTA
10 mM NaCl
Dissolve all salts in deionized water to the final desired concentrations
Bring pH to 8.0 with NaOH - Column elution buffer
50 mM EPPS-NaOH, pH 8.0
1mM EDTA
0.8 M NaCl
Dissolve all salts in deionized water to the final desired concentrations
Bring pH to 8.0 with NaOH - Specificity (SC/O) buffer
30 mM triethanolamine
15 mM magnesium acetate, pH 8.3
Dissolve all salts in deionized water to the final desired concentrations
Bring pH to 8.3 with acetic acid
Store at 4 °C - Assay buffer (1 ml)
30 mM triethanolamine
15 mM magnesium acetate, pH 8.3
10 μg ml-1 carbonic anhydrase - 4x SDS reducing buffer for SDS-PAGE (Optional)
125 mM Tris-HCl pH 6.8
4% (w/v) SDS
0.01% (w/v) bromophenol blue
20% (v/v) glycerol
75 mM 2-mercaptoethanol
Dissolve all salts in deionized water to the final desired concentrations
Add glycerol and 2-mercaptoethanol before use - BS buffer for SDS-PAGE (Optional)
10 mM Tris-HCl, pH 7.5
150 mM NaCl
Dissolve all salts in deionized water to the final desired concentrations
Acknowledgments
We thank the reviewers for helpful comments that improved the article. Funding for J. N.Y. was through ANU visiting scholar (CE140100015) and NSF Grant 1040965. A.H. was funded through a Clarendon Scholarship, Oxford and ANU visiting scholar (CE140100015). R.E.S was funded through ARC DECRA scheme (DE13010760). R.E.M.R. was funded through ERC Starting Grant (SP2-GA-2008-200915), F.M.M.M. was funded through NSF Grant 104095 and S.M.W was funded through Australian Research Council Grant CE14010001. We would like to thank Bratati Mukherjee for assistance in the video of the French Press.
References
- Andrews, T. J. (1988). Catalysis by cyanobacterial ribulose-bisphosphate carboxylase large subunits in the complete absence of small subunits. J Biol Chem 263(25): 12213-12219.
- Falkowski, P. G. and Raven, J. A. (2007). Aquatic photosynthesis. Princeton University Press.
- Galmes, J., Kapralov, M. V., Andralojc, P. J., Conesa, M. A., Keys, A. J., Parry, M. A. and Flexas, J. (2014). Expanding knowledge of the Rubisco kinetics variability in plant species: environmental and evolutionary trends. Plant Cell Environ 37(9): 1989-2001.
- Kane, H. J., Viil, J., Entsch, B., Paul, K., Morell, M. K. and Andrews, T. J. (1994). An improved method for measuring the CO2/O2 specificity of ribulosebisphosphate carboxylase-oxygenase. Aust J Plant Physiol 21(4): 449-461.
- Kane, H. J., Wilkin, J. M., Portis, A. R. and John Andrews, T. (1998). Potent inhibition of ribulose-bisphosphate carboxylase by an oxidized impurity in ribulose-1,5-bisphosphate. Plant Physiol 117(3): 1059-1069.
- Losh, J. L., Young, J. N. and Morel, F. M. M. (2013). Rubisco is a small fraction of total protein in marine phytoplankton. New Phytol 198(1): 52-58.
- Sharwood, R. E., Ghannoum, O., Kapralov M. V., Gunn L. H. and Whitney, S. M. (2016). Variation in response of C3 and C4 Paniceae Rubisco to temperature provides opportunities for improving C3-photosynthesis. Nat Plants 2: 16186
- Sharwood, R. E., von Caemmerer, S., Maliga, P. and Whitney, S. M. (2008). The catalytic properties of hybrid Rubisco comprising tobacco small and sunflower large subunits mirror the kinetically equivalent source Rubiscos and can support tobacco growth. Plant Physiol 146(1): 83-96.
- Sunda, W. G., Price, N. M. and Morel, F. M. M. (2005). Trace metal ion buffers and their use in culture studies. In: Anderson, R. A. (Ed.). Algal culturing techniques. Elevier Academic Press, pp: 35-63.
- Tcherkez, G. G., Farquhar, G. D. and Andrews, T. J. (2006). Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc Natl Acad Sci U S A 103(19): 7246-7251.
- Whitney, S. M., Baldet, P., Hudson, G. S. and Andrews, T. J. (2001). Form I Rubiscos from non-green algae are expressed abundantly but not assembled in tobacco chloroplasts. Plant J 26(5): 535-547.
- Whitney, S. M. and Sharwood, R. E. (2014). Plastid transformation for Rubisco engineering and protocols for assessing expression. In Maliga, P. (Ed.). Chloroplast Biotechnology. Humana Press, pp 245-262.
- Whitney, S. M., Houtz, R. L. and Alonso, H. (2011). Advancing our understanding and capacity to engineer nature’s CO2-sequestering enzyme, Rubisco. Plant Physiol 155(1): 27-35.
- Whitney, S. M. and Sharwood, R. E. (2007). Linked Rubisco subunits can assemble into functional oligomers without impeding catalytic performance. J Biol Chem 282(6): 3809-3818.
- Young, J. N., Heureux, A. M., Sharwood, R. E., Rickaby, R. E., Morel, F. M. M. and Whitney, S. M. (2016). Large variation in the Rubisco kinetics of diatoms reveals diversity among their carbon-concentrating mechanisms. J Exp Bot 67(11): 3445-3456.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Young, J. N., Heureux, A. M. C., Rickaby, R. E. M., Morel, F. M. M., Whitney, S. M. and Sharwood, R. E. (2017). Rubisco Extraction and Purification from Diatoms. Bio-protocol 7(6): e2191. DOI: 10.21769/BioProtoc.2191.
Category
Plant Science > Plant physiology > Photosynthesis
Biochemistry > Other compound > Chlorophyll
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











