- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Automatic Quantification of the Number of Intracellular Compartments in Arabidopsis thaliana Root Cells
Published: Vol 7, Iss 4, Feb 20, 2017 DOI: 10.21769/BioProtoc.2145 Views: 13461
Reviewed by: Tie LiuAmey G RedkarHonghong Wu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2371 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1723 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 791 Views
Abstract
In the era of quantitative biology, it is increasingly required to quantify confocal microscopy images. If possible, quantification should be performed in an automatic way, in order to avoid bias from the experimenter, to allow the quantification of a large number of samples, and to increase reproducibility between laboratories. In this protocol, we describe procedures for automatic counting of the number of intracellular compartments in Arabidopsis root cells, which can be used for example to study endocytosis or secretory trafficking pathways and to compare membrane organization between different genotypes or treatments. While developed for Arabidopsis roots, this method can be used on other tissues, cell types and plant species.
Keywords: ArabidopsisBackground
Studies on plant intracellular trafficking have widely beneficiated from the identification and characterization of proteins that are localized to specific intracellular compartments. These proteins can serve in subsequent studies as compartment markers either using immunofluorescence when antibodies are available, or direct fusion with fluorescent proteins (Dettmer et al., 2006; Geldner et al., 2003; Jaillais et al., 2008; Jaillais et al., 2006). Typically, they can be used in co-localization experiments (Geldner et al., 2009; Simon et al., 2014), but also as reference points to characterize mutants, drugs or growth conditions that might affect intracellular trafficking pathways, such as exocytosis, endocytosis, autophagy, or secretory trafficking. The localization of these marker proteins may vary in different manner, including for example their number, size, shape, clustering or labeling intensity. For example, in root, the fungal toxin Brefeldin A (BFA), an inhibitor of protein recycling, degradation and secretion, induces the aggregation of multiple compartments in or around the so-called ‘BFA compartment’ (Geldner et al., 2003; Geldner et al., 2009). Wortmannin, an inhibitor of PI3 Kinase activity induces the fusion of late endosomal compartments (Jaillais et al., 2006; Tse et al., 2004), while concanamycin A induces TGN swelling (Dettmer et al., 2006). Accordingly similar effects on compartment numbers and/or morphology have been observed in trafficking mutants (Dettmer et al., 2006; Geldner et al., 2003; Jaillais et al., 2007; Sauer et al., 2013).
Automatic spot detection was spearheaded on leaf for the detection of endomembrane rearrangements induced by environmental changes such as dark, cold treatment or biotic stresses (Salomon et al., 2010). This technique was also applied to study the endocytosis of the FLAGELIN-INSENSITIVE2 (FLS2) receptor (Beck et al., 2012; Mbengue et al., 2016; Spallek et al., 2013). Here we described a protocol to computationally detect and count the number of intracellular compartments on Arabidopsis root image. This protocol relies on a macro that runs on the open source image analysis software ImageJ and that can work with a wide variety of images with different image-to-noise signals. In addition, it proposes two different modes of detection; a first one where the macro automatically finds the root area and another one that allows the selection of a user-defined region of interest (ROI). Finally, although this version of the macro is designed to count the number of spots, similar image segmentation can easily be used to measure spot size, to estimate signal intensity, to capture compartment morphology or to automatically quantify co-localization between two or more channels.
Equipment
- Microscope: Plant imaging was performed on an inverted Zeiss microscope (Zeiss, model: AxioObserver Z1 ) equipped with a spinning disk module (Yokogawa, model: CSU-W1-T3 )
- Camera: ProEM+ 1024B camera (Princeton Instrument, model: ProEM+ 1024B)
- Objective: 63x Plan-Apochromat objective (numerical aperture 1.4, oil immersion).
Note: Images may be performed with any confocal microscope. We describe above the microscope setup used to take the sample images that can be downloaded below (see Software 7).
Software
- ImageJ (http://imagej.nih.gov/ij/) (Schneider et al., 2012) (see Note 1)
Note: If link does not open, copy-paste the address in your browser. - SiCE spot detector Macro for ImageJ
Note: Download SiCE spot detector Macro for ImageJ. Go to http://www.ens-lyon.fr/RDP/SiCE/METHODS.html, right click on SiCE ‘SpotDetector.ijm’ and choose ‘save link as’. Use the following name to save the file: ‘SiCE SpotDetectorV3.ijm’. - Wavelet A Trou plugin for ImageJ (http://www.ens-lyon.fr/RDP/SiCE/METHODS_files/Wavelet_A_Trou.jar)
Note: If link does not open, copy-paste the address in your browser. - FeatureJ plugin for ImageJ (http://www.imagescience.org/meijering/software/featurej/)
Note: If link does not open, copy-paste the address in your browser. - XLstat (Addinsoft, https://www.xlstat.com).
- R (The R foundation, https://www.r-project.org/), Excel (Microsoft, https://products.office.com/).
- (Optional) Download template images (http://www.ens-lyon.fr/RDP/SiCE/METHODS) (see Note 2).
Procedure
- Images acquisition
The macro described below can work with any type of confocal images whether taken with a laser scanning confocal microscope or with a spinning disk confocal microscope. You should therefore define acquisition parameter according to your microscope set-up. Note that for quantitative imaging, pictures should be taken with detector settings optimized for low background and no pixel saturation (Simon et al., 2016). For comparison purposes, we recommend using similar confocal settings for all images. Exposure time must be defined by taking into account compartments movement during acquisition. Too long exposure might lead to abnormal shape or multiple counts of the same intracellular particles.
As an example, using the spinning disk microscope described in the equipment section above, we typically use the following settings: image acquisition time 200 msec, laser power 30%, EM gain 100 and image size 1024 x 1024 pixels.
Note: The settings used might vary greatly depending on the microscope set-up and the strength of the marker line. - Plugin installation (Video 1)
- Copy Wavelet_A_Trou file and FeatureJ plugins in ImageJ/plugins folder.
- Copy SiCE Spot DetectorV3.ijm file in ImageJ/macros folder.
- Install the SiCE spot detector macro by selecting it in Plugins > Macros > Install. SiCE SpotDetectorV3 should appear in the Plugins > Macros > lower panel. Video 1. Procedure to install the Sice Spotdetector Macro and related Plugins
- Copy Wavelet_A_Trou file and FeatureJ plugins in ImageJ/plugins folder.
- Setting up parameters for single image analysis
The purpose of single image analysis (Procedure C) is to find the best parameters possible to segment your image correctly (i.e., best parameters to find all [or most] of the compartments in the image with as little false positive as possible) and then use these parameters in batch mode (Procedure D). Every marker lines have different signal strength, particle size and signal-to-noise ratio. It is impossible to predict which will be the best parameters to use for a specific marker line. Therefore, these parameters have to be determined empirically for each marker lines. The procedure in C allows testing a range of parameters to confront the segmentation results with your input image (step C10). Mostly, the use of these parameters are empirical and we advise you to explore different ones in order to find the best setting for your image (below, we provide some numbers that we find often work well for counting endosome numbers in Arabidopsis root). However some of these parameters are directly dependent on the type of particles you want to analyze, notably the size and circularity. Particle size can be estimated using the optional step C3, but you can also try different size parameters empirically as well and by-pass step C3.- Open a confocal image with ImageJ (or Fiji) software in which you want to quantify the number of intracellular compartments or template pictures (see Note 2). It is possible to use a projected 3D-stack. However, we do not recommend using projections that might yield artifacts and/or mask the presence of some particles and we recommend instead quantifying all the images in the Z-stack using the batch mode (see Procedure D).
- If you want to use scaled images, ‘Set Scale’ in the ‘Analyze Tab’ according to your microscope set-up, otherwise proceeds directly to step C3.
- Estimate particle size with ImageJ (Optional STEP, Video 2). Use straight-line tool to draw a line through several compartments. Then, use the plot profile tool (Analyze > Plot profile) and measure the size of the structures within the profile by using again the straight-line tool. Measure several compartments small and big ones approximately at the base of the peaks (Figure 1). It will help you to determine maximum size, minimum size and sigma parameters, which will be useful for detection step (see below step C8). As an example, see Figure 2, which shows the typical size of various compartments obtained with the microscope set-up described above (see Equipment section).
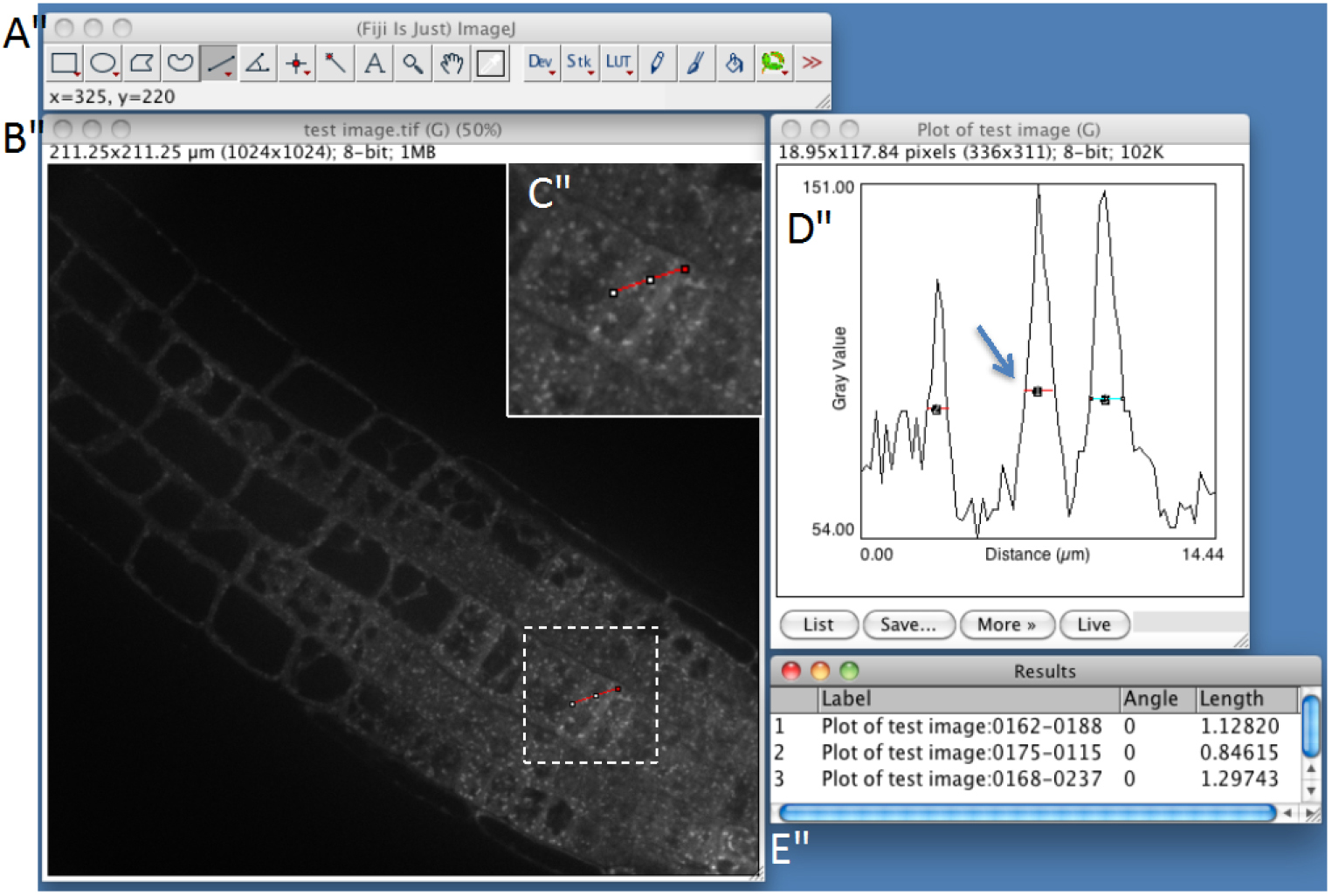
Figure 1. Snapshot from FIJI program showing manual measurement of individual spot-like structures using plot-profile and straight-line tools (step C3). A. FIJI main toolbar; B. Test image window; C. Enlargement of the dashed-square in (B), showing the red line that is drawn across several compartments using the straight-line tool of ImageJ. D. Fluorescence plot along the red line. Compartments are identified as peaks and are measured by drawing lines at their base. E. Peak width measurements.Video 2. Procedure to estimate the size of the compartments of interest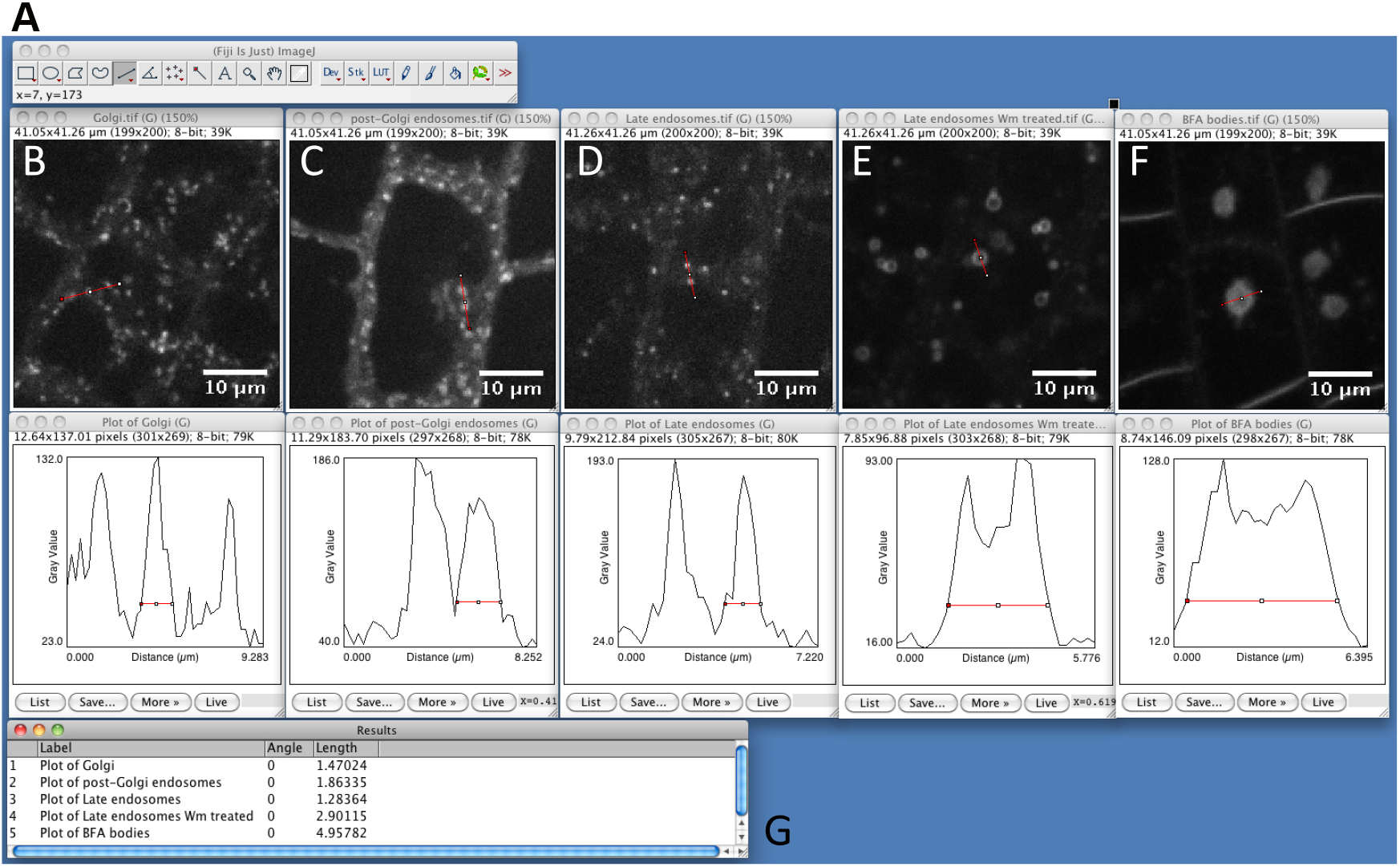
Figure 2. Snapshot from ImageJ program showing the size of different intracellular compartments as captured by spinning disk microscopy. A. FIJI main toolbar; B. Golgi apparatus, marker line is Wave18 (W18) (Geldner et al., 2009). C. Post-Golgi endosomal compartments, marker line is Wave25 (W25) (Geldner et al., 2009). D. Late endosomal compartments, marker line is Wave7 (W7) (Geldner et al., 2009). E. Wortmannin compartments. Late endosomes (W7) fuse into the so-called ‘Wortmannin compartment’ following treatment with the PI3Kinase/PI4Kinase inhibitor Wortmannin (Wm, 30 µM for 60 min) (Jaillais et al., 2006; Simon et al., 2014; Simon et al., 2016). F. BFA compartments. Endosomes labeled by the endocytic tracer FM4-64 aggregates into the so-called ‘BFA compartment’ following treatment with the fungal toxin BrefeldinA (BFA, 50 µM for 60 min) (Geldner et al., 2003; Geldner et al., 2009). G. Summary table of peak width measurements for the marker shown in A to F. - Launch SICE-Spot Detector Macro from Plugins > Macro > SpotDetectorMacroV3 (Video 3). Video 3. Procedure to find the best parameters to segment intracellular particles with the SiCE spot detector macro
- Uncheck Batch mode to start single image analysis.
- Select if you want to use scaled images or un-scaled images, by checking/unchecking the ‘scaled image’ box. Scaled image will provide results expressed in number of spots/µm2, while un-scalded images will provide result expressed in number of spot/pixel2
- Uncheck Automatic ROI if you want to manually draw region of interest (Note 3).
- Select parameters (Figure 2)
First three parameters directly arise from step C3. Min Particle Size, Max Particle Size and Circularity will be used in the Built-in Macro Function Analyse Particles (https://imagej.nih.gov/ij/docs/guide/146-30.html). Particles with size outside the range defined are ignored. Values are expressed either in square µm or square pixels. Circularity depends on the shape of the compartments and range from 0 (infinitely elongated polygon) to 1 (perfect circle). Classically, intracellular compartments in Arabidopsis roots, such as endosomes, can be detected using 0.5-5 µm2 and 0.5-1 circularity (but other parameters may be used to quantify more elongated particles, or bigger particles such as BFA compartments).- In confocal images Intracellular compartments of interest will appear as more or less round shaped objects with higher fluorescent signal in the center. Image convolution by a Gaussian kernel is generally sufficient for segmenting them. Sigma parameter corresponds to standard deviation of the distribution used in the Gaussian filtering method (Lowe, 2004). Adjusting Sigma will depend on the background within cells and your object size and has to be determined empirically.
- The Gaussian filtering that will be applied to your image can be chosen from Difference of Gaussian ‘DoG’ or Laplacian of Gaussian. Classically, Laplacian of Gaussian filtering gives the sharpest boundaries of the objects allowing size measurement, while DoG filtering will be more efficient in findings structures of different sizes or in a higher background.
- Auto-threshold corresponds to the method/algorithm that will be applied to your filtered image (Triangle, Otsu and Huang) (see more details about this step at the following address http://imagej.net/Auto_Threshold). Again, you need to try these different methods to find the best one empirically.
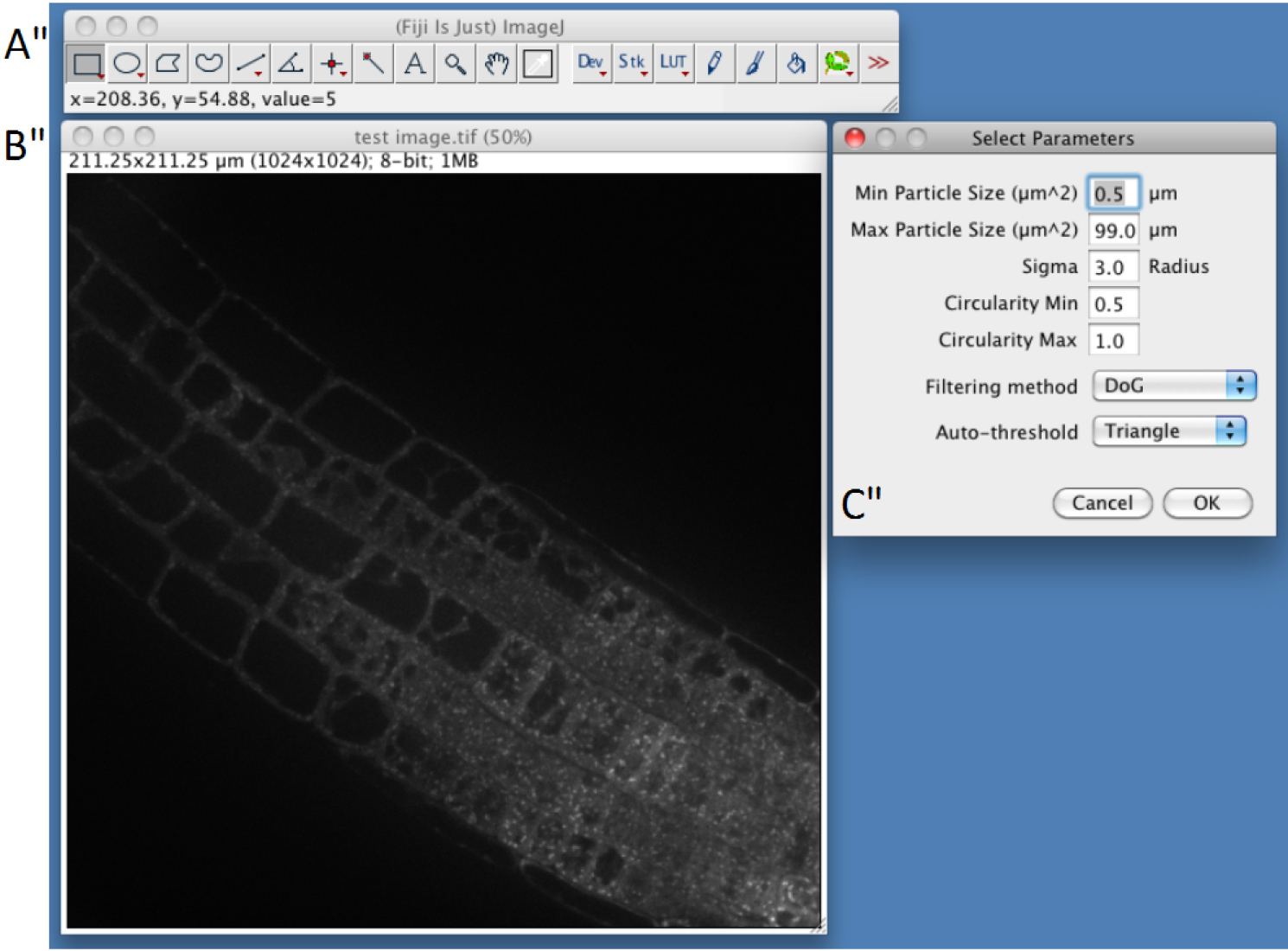
Figure 3. Snapshot from ImageJ program showing selection of parameters step. A. FIJI main toolbar; B. Test image window; C. Select Parameters selection windows of the SICE spot detector.
- In confocal images Intracellular compartments of interest will appear as more or less round shaped objects with higher fluorescent signal in the center. Image convolution by a Gaussian kernel is generally sufficient for segmenting them. Sigma parameter corresponds to standard deviation of the distribution used in the Gaussian filtering method (Lowe, 2004). Adjusting Sigma will depend on the background within cells and your object size and has to be determined empirically.
- Once you have chosen all your parameters, click ‘OK’ to proceed with the analysis.
- If you chose manual mode type OK and define a Region of Interest (ROI). Compartments within the defined area will be segmented according to the parameters, methods and modes chosen previously in step C8.
- If you chose automatic ROI (step C7), the macro will automatically define the portion of the image that corresponds to the root tissue with the highest signal (see red line in Figure 4) (see Note 3) and will discard the rest of the image for subsequent analyses.
- A merge image between segmented compartments boundaries and original fluorescent image will be displayed to evaluate the efficiency of the process (Figure 4); i.e., it will help you to evaluate if the parameters you used are the best to find the intracellular compartments of interest. ROI will be displayed on the original image as a red surrounded region. By clicking ‘OK’ you can return to step C8 to modify parameters and make new analyses.
- In manual ROI selection mode, you will be able to test a new set of parameters on the same manually defined ROI (ROI defined previously in step C9a). You will also have the option to define a new ROI.
- Automatic ROI allows you to switch from automatic to manual ROI selection of vice-versa.
- Reiterate steps C6-C10 until you find the best parameters to segment intracellular compartments in your image and then proceed to step C12.
- Uncheck the ‘unhappy’ box to display a ‘Summary’ table with ‘Picture name’, ‘Cell Area’, ‘Spot number’ and parameters used. Once you determined optimum parameters for your data set, you can relaunch the macro for batch mode analysis (see Procedure D).
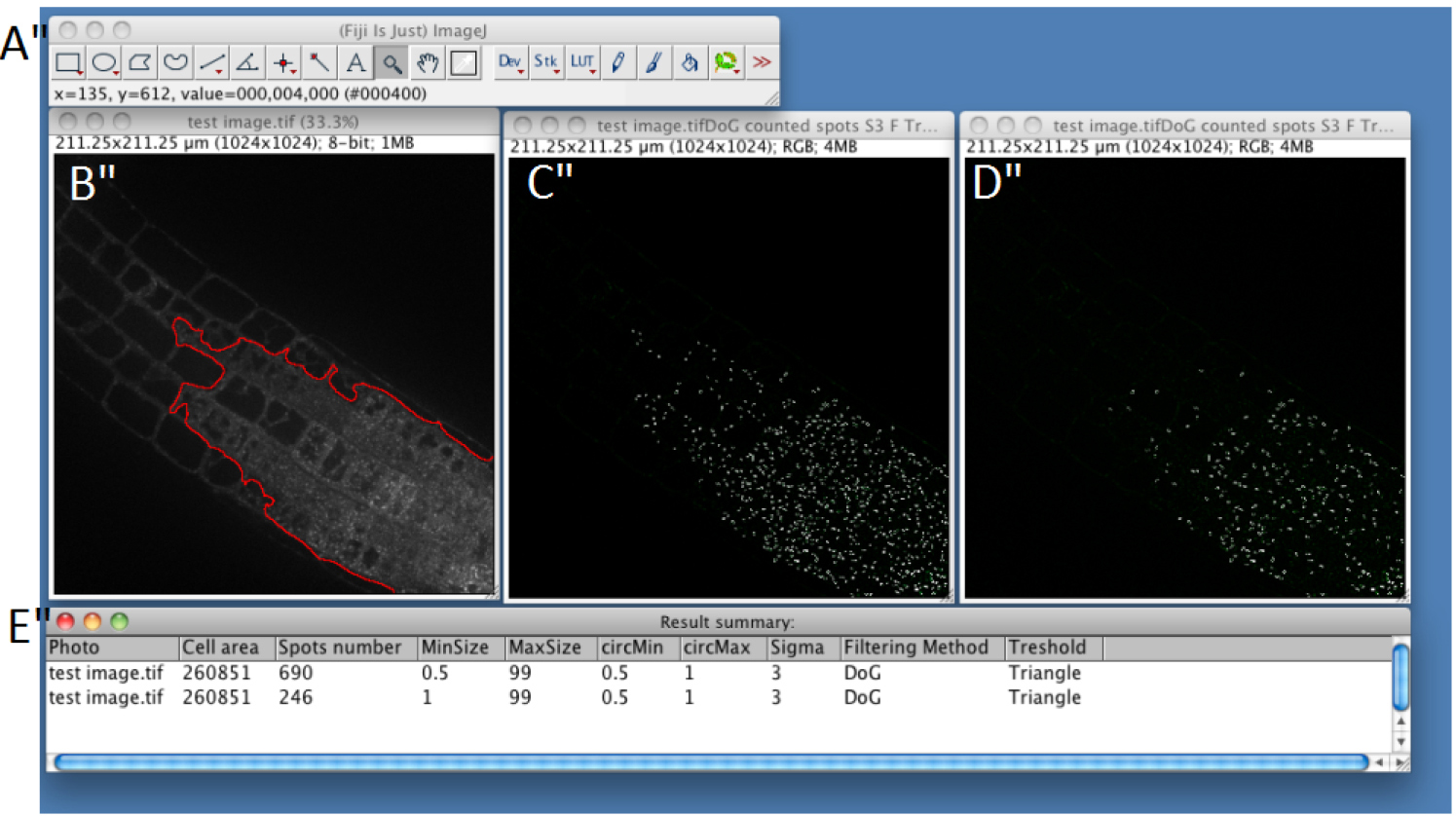
Figure 4. Snapshot from ImageJ program showing the results for single image analysis. A. FIJI main toolbar; B. Test image window. Region of Interest is represented as a red line. C. Result of spot detection using the following parameters: MinSize 0.5 MaxSize 99 circMin 0.5 circMax 1 Sigma 3 Filtering Method Difference of Gaussian Threshold Triangle; D. Result of spot detection using the following parameters: MinSize 1 MaxSize 99 circMin 0.5 circMax 1 Sigma 3 Filtering Method Difference of Gaussian Threshold Triangle; D. ‘Result summary’ table showing the quantification of the number of spot in the different condition tested, using the parameters defined as in C for line 1 and as in D for line 2.
- Open a confocal image with ImageJ (or Fiji) software in which you want to quantify the number of intracellular compartments or template pictures (see Note 2). It is possible to use a projected 3D-stack. However, we do not recommend using projections that might yield artifacts and/or mask the presence of some particles and we recommend instead quantifying all the images in the Z-stack using the batch mode (see Procedure D).
- Batch images analysis
- Launch SICE-Spot Detector Macro from Plugins > Macro > SpotDetectorMacroV3.
- Choose Source directory.
- Choose Destination directory where the merged images will be saved (see step C10) (these files allow you to check that segmentation was correctly performed in all images).
- Select the previously optimized parameters that will be used for analyzing your images.
- Macro will perform Spot detection on all images contained in the Source directory and collect results in the Summary table. This table can be directly saved as an Excel file or copied and pasted in a pre-existing table.
- Launch SICE-Spot Detector Macro from Plugins > Macro > SpotDetectorMacroV3.
Data analysis
When manually counting the number of spots per cells, 100 cells are typically counted on 10 to 20 different roots (Simon et al., 2016). We suggest keeping a similar number of independent roots (10 to 20) when using automatic spot detection, which will ultimately represent around 500 to 1,000 cells. However, because variability may vary according to the type of spots and markers used, we advise to first estimate sample size. For this purpose, quantify the number of spots on a small numbers of samples and estimate sample size using the following formula:
n = (t x t) x (s x s)/(d x d)
Where,
s is standard error of the mean,
t is the statistical confidence level (1.96 as deduced from the standard normal distribution for a confidence interval of 95%),
d is the margin of error (0.05).
Biological triplicates are suggested. For statistical analyses, choose parametric or non-parametric test depending on sample distribution (Gaussian distribution or not, respectively). Parametric test such as Tukey’s honestly significant difference test or non parametric test such as Kruskal-wallis test or Dwass-Steel-Critchlow-Fligner procedure for multiple comparison between samples can be performed with XLstat software (http://www.xlstat.com/) or R software (https://www.r-project.org/), in order to find if means are significantly different from each other. Graphic representation can be done using either R software or Excel software (Microsoft, https://products.office.com/).
Notes
- The SiCE SpotDetector macro works with the current distribution of ImageJ (as of November 2016). The only required plugins are Wavelet_A_Trou and FeatureJ. If you have an older version of ImageJ and encounter problems with the macro, re-install the newest version of ImageJ (http://imagej.nih.gov/ij/).
- To test the macro, you can download sample images (http://www.ens-lyon.fr/RDP/SiCE/METHODS) (set scale at distance in pixels: 4.8473, known distance: 1, pixel aspect ratio 1.0, Unit of length: µm).
- This method will automatically detect the root in your picture using wavelet-based image segmentation. It will either select the entire root in the image, or the part of the root that has the highest signal. The macro selects only one continuous area per image. This will allow the macro to quantify compartments only in the area with highest signal in the case of uneven labeling along the root. If you want to quantify several areas in the same root with significant difference in labeling intensity, crop your pictures according to these different areas or use the normal mode and manually define ROI.
Acknowledgments
We thank Laia Armengot (RDP, ENS Lyon) for critical comments on the manuscript, Erik Meijering (Erasmus University Medical Center, Rotterdam, The Netherlands) for developing FeatureJ. Y.J. is funded by ERC No. 3363360-APPL under FP/2007-2013.
References
- Beck, M., Zhou, J., Faulkner, C., MacLean, D. and Robatzek, S. (2012). Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24(10): 4205-4219.
- Dettmer, J., Hong-Hermesdorf, A., Stierhof, Y. D. and Schumacher, K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18(3): 715-730.
- Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A. and Jurgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112(2): 219-230.
- Geldner, N., Denervaud-Tendon, V., Hyman, D. L., Mayer, U., Stierhof, Y. D. and Chory, J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59(1): 169-178.
- Jaillais, Y., Fobis-Loisy, I., Miege, C. and Gaude, T. (2008). Evidence for a sorting endosome in Arabidopsis root cells. Plant J 53(2): 237-247.
- Jaillais, Y., Fobis-Loisy, I., Miege, C., Rollin, C. and Gaude, T. (2006). AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443(7107): 106-109.
- Jaillais, Y., Santambrogio, M., Rozier, F., Fobis-Loisy, I., Miege, C. and Gaude, T. (2007). The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell 130(6): 1057-1070.
- Lowe, D. G. (2004). Distinctive image features from scale-invariant keypoints. Int J Comput Vision 60(2): 91-110.
- Mbengue, M., Bourdais, G., Gervasi, F., Beck, M., Zhou, J., Spallek, T., Bartels, S., Boller, T., Ueda, T., Kuhn, H. and Robatzek, S. (2016). Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc Natl Acad Sci U S A 113(39): 11034-11039.
- Salomon, S., Grunewald, D., Stuber, K., Schaaf, S., MacLean, D., Schulze-Lefert, P. and Robatzek, S. (2010). High-throughput confocal imaging of intact live tissue enables quantification of membrane trafficking in Arabidopsis. Plant Physiol 154(3): 1096-1104.
- Sauer, M., Delgadillo, M. O., Zouhar, J., Reynolds, G. D., Pennington, J. G., Jiang, L., Liljegren, S. J., Stierhof, Y. D., De Jaeger, G., Otegui, M. S., Bednarek, S. Y. and Rojo, E. (2013). MTV1 and MTV4 encode plant-specific ENTH and ARF GAP proteins that mediate clathrin-dependent trafficking of vacuolar cargo from the trans-Golgi network. Plant Cell 25(6): 2217-2235.
- Schneider, C. A., Rasband, W. S. and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7): 671-675.
- Simon, M. L., Platre, M. P., Assil, S., van Wijk, R., Chen, W. Y., Chory, J., Dreux, M., Munnik, T. and Jaillais, Y. (2014). A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant J 77(2): 322-337.
- Simon, M. L., Platre, M. P., Marques-Bueno, M. M., Armengot, L., Stanislas, T., Bayle, V., Caillaud, M. C. and Jaillais, Y. (2016). A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat Plants 2: 16089.
- Spallek, T., Beck, M., Ben Khaled, S., Salomon, S., Bourdais, G., Schellmann, S. and Robatzek, S. (2013). ESCRT-I mediates FLS2 endosomal sorting and plant immunity. PLoS Genet 9(12): e1004035.
- Tse, Y. C., Mo, B., Hillmer, S., Zhao, M., Lo, S. W., Robinson, D. G. and Jiang, L. (2004). Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16(3): 672-693.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Bayle, V., Platre, M. P. and Jaillais, Y. (2017). Automatic Quantification of the Number of Intracellular Compartments in Arabidopsis thaliana Root Cells. Bio-protocol 7(4): e2145. DOI: 10.21769/BioProtoc.2145.
Category
Plant Science > Plant cell biology > Cell imaging
Cell Biology > Cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












