- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Transformation of Thermus Species by Natural Competence
Published: Vol 6, Iss 22, Nov 20, 2016 DOI: 10.21769/BioProtoc.2007 Views: 11074
Reviewed by: Daan C. SwartsMaria SinetovaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Fast and Easy Method to Study Ralstonia solanacearum Virulence upon Transient Gene Expression or Gene Silencing in Nicotiana benthamiana Leaves
Wenjia Yu and Alberto P. Macho
Aug 5, 2021 4701 Views

A New Tool for the Flexible Genetic Manipulation of Geobacillus kaustophilus
Ryotaro Amatsu [...] Ken-ichi Yoshida
Sep 5, 2022 1905 Views

Efficient Genetic Transformation and Suicide Plasmid-mediated Genome Editing System for Non-model Microorganism Erwinia persicina
Tingfeng Cheng [...] Lei Zhao
Mar 20, 2024 2443 Views
Abstract
Many Thermus species harbour genomes scourged with horizontally transferred signatures. Thermus thermophilus (Tth) has been characterized as naturally competent. The transformation protocol described here is based on the maximum DNA uptake rates registered at mid-exponential phase (OD600 0.3-0.4). Here we describe the stepwise protocol followed for transformation of both plasmids and linearized genomic DNA, of which the latter can be employed as an alternative method to electroporation to introduce mutations or to generate gene deletions in Thermus isolates, for instance.
Background
Thermus thermophilus (Tth) is an extreme thermophilic species extensively used as laboratory model, due to its ancient phylogenetic origin, the comparative ease of crystallisation of its proteins and macromolecular complexes, and the fast growth and good yields under laboratory growth conditions of several of its isolates. Among the most commonly employed isolates, the strains T. thermophilus HB27 and HB8 constitute the most frequently used models due to the high rates by which they can be transformed by natural competence. DNA of any source and topology can be easily taken up by growing cells of these isolates at rates of around 40 kb/s/cell (Schwarzenlander and Averhoff, 2006) showing yields of around 10-2 transformants/viable cell. For this, a polar located natural competence apparatus (Gold et al., 2015) involving at least 16 proteins encoded in five loci in the chromosome (Averhoff, 2009) of both strains is expressed essentially in a constitutive way, although the transformation efficiency is higher at exponential phase. Variants of antibiotic resistance genes encoding thermostable variants have been developed by directed evolution as gene markers, in such a way that selection can be performed in plates with kanamycin (Lasa et al., 1992), hygromycin B (Nakamura et al., 2005), or bleomycin (Brouns et al., 2005). Streptomycin resistance can be also employed to check natural competence with isogenic DNA (Koyama et al., 1986). This protocol describes the highly efficient transformation of cultures at exponential growth phase, providing reproducible data of maximized transformation efficiency.
Materials and Reagents
- 12 ml sterile plastic tubes
- 15 ml sterile plastic tubes
- Sterile plastic Petri dish plates (standard size; 100 x15 mm)
- Sterile tips for micropippettes, 5-200 µl (for instance, Daslab, catalog number: 162001 )
- Sterile tips for micropippettes, 100-1,000 µl (for instance, Daslab, catalog number: 162222 )
- Sterile tips for micropippettes, 10 µl (for instance, Metler Toledo, catalog number: 17004280 )
- Recipient Thermus strain, for instance, HB27 wild type (HB27/ATCC BAA-163 /DSM 7039)#
#Note: Other Thermus thermophilus strains such as NAR1 and HB8, have been defined as naturally competent, thus, harboring the competence proteins required for transformation (César et al., 2011). All Thermus spp. strains referred here are available at the DSMZ collection. - Kanamycin sulphate (Sigma-Aldrich, catalog number: K1377-5G )
- Tryptone (Conda, catalog number: 1612 )
- Yeast extract (Conda, catalog number: 1702 )
- Sodium chloride (NaCl) (EMD Millipore, catalog number: 1.06404.1000 )
- Agar (grade A) (Conda, catalog number: 1800 )
- Thermus water (carbonated-rich mineral water)
- DNA template: purified plasmids or genomic DNA** (harbouring the thermostable kat cassette)
**Note: DNA quantity depends on the topology of the DNA to be transformed and the competence of the recipient strain. - TB liquid medium (see Recipes)
- TB agar plates (see Recipes)
Equipment
- 50 ml Erlenmeyer flask
- Rotary shaker incubator reaching 70 °C (150 rpm) (Thermo Scientific, Thermo ScientificTM, catalog number: SHKE 420HP )
- Glass beads or a spreader
- Autoclave device (CertoClav, model: EL 18L )
- Spectrophotometer (Hitachi, model: U-2000 )
- Pipette P2, 0.2-2 μl (Gilson, Pipetman classicTM, catalog number: F144801 )
- Pipette P20, 2-20 μl (Gilson, Pipetman classicTM, catalog number: F123600 )
- Pipette P100, 20-100 μl (Gilson, Pipetman classicTM, catalog number: F123615 )
- Pipette P200, 50-200 μl (Gilson, Pipetman classicTM, catalog number: F123601 )
- 70 °C heater (Gemini, model: Memmert UE400 )
- Humid chambers
Note: Tupperware devices are preferentially used. Humidified plastic bags are also valid. Addition of a paper towel, soaked on Thermus water and laid on the bottom of the chamber would enhance humidity maintenance. Ventilation should be avoided in order to conserve humidity. An example is provided in the photographs beneath (Figure 1).
Figure 1. Humid chambers. Descriptive photographs of the humid chambers used to incubate Thermus plates. Place a piece of paper soaked with Thermus water and a bottle cap filled with the same water on the bottom of the chambers to maintain high humidity.
Procedure
- Culture preparation: growth of the subject strain to be transformed (see Figure 2).
- Inoculate 50 μl of saturated liquid growth culture (optical density at 600 nm [OD600] aprox. 2) in 10 ml TB medium in a 50-ml Erlenmeyer or inoculate the culture from the glycerol stock or from a frozen pellet. In the latter cases, one additional day growth would be required to reach optimal culture fitness.
- Incubate overnight the culture at 65 °C in a shaker incubator at 150 rpm.
- Dilute the overgrown culture in 10 ml fresh TB medium to OD600 = 0.05 in a 50-ml Erlenmeyer.
Notes: - If the culture has not reached saturation after overnight growth, postposition of the experiment is highly recommended.
- If the culture has not grown optimally overnight and the experiment should be performed, a starting OD600 of 0.07-0.08 is possible.
- Incubate in a shaker incubator at 65 °C until an OD600 of 0.3-0.4 is reached.
Note: An OD600 close to 0.3 is recommended. Figure 2. Schematic representation of the Thermus transformation procedure. Plate shows a dilution of a transformation of 100 ng of a linearized plasmid harbouring a kat cassette flanked by two genomic arms for double recombination in HB27 strain.
Figure 2. Schematic representation of the Thermus transformation procedure. Plate shows a dilution of a transformation of 100 ng of a linearized plasmid harbouring a kat cassette flanked by two genomic arms for double recombination in HB27 strain. - DNA addition
- Add 0.01-1 μg of DNA (genomic ~0.01 μg; plasmids ~0.1-0.25 μg; PCR products ~1 μg) in a sterile 12 ml plastic tube. Tubes can be pre-heated at 65 °C.
Notes: - Genomic DNA (carrying a selectable marker integrated in the genome) should not be added in higher quantities to avoid recombination farther than the target.
- DNA samples should be purified following standard protocols and eluted in water or TE.
- Pre-heating of the tubes keeps the temperature of the growing culture stable.
- Add 500 μl of the exponentially growing culture (see Figure 2) to the DNA.
- As a control, prepare 15 ml plastic tube without DNA, and add 500 μl of the exponentially growing culture to this tube.
- DNA transformation
- Incubate each tube in a shaker incubator at 65 °C for 4 h.
- Plate 50 μl and 400 μl of the cells on selective agar medium, spread by using glass beads or a spreader.
Notes: - Volume plated depends on the competence efficiency of the employed Thermus strain. Previous assays plating serial dilution spots can shed light on the appropriate volume to plate.
- Plate the control tube like the others, in addition to plating on a non-selective agar medium, as it will work as the reference of viable cells.
- Incubate plates in humid chambers for 48 h at 65 °C or until colonies become visible.
Notes: - Ordinarily, incubation temperature is set according to optimal growth of the strain but it should be adapted to the limitations of the selectable marker employed in the transformations too (for instance, hph5 conferring hygromycin resistance cannot be exposed to temperatures higher than 60 °C whereas kanamycin is selectable in temperatures higher than 70 °C).
- Transformation yields highly depend on the DNA template employed and the strain being transformed. For instance, HB27 transformed with 100 ng of plasmidic DNA shows rates of 10-3 transformants per viable cell.
Data analysis
- For transformation assays, a set of statistical tests should be performed to compare and analyse the transfer frequencies obtained.
- Transfer frequencies are calculated as the number of CFU transformants divided by the number of CFU of viable cells. To examine differences among transfer frequencies, Student’s t-test and Mann-Whitney U-tests may be used. When comparing transformability of a strain testing several DNA templates, examination of the differences among and within various DNA templates transfer frequencies can be addressed by one-way ANOVA. If different strains are tested, Wilcoxon-tests may be used to evaluate differences among their transfer frequencies. Simple linear regression test can be implemented for modelling the relationship between the DNA template and the frequency of transfer. Post-hoc Tukey and Bonferroni tests should be applied when convenient.
- At least, 3 independent experiments, should be performed for reliable and reproducible data.
Notes
- This transformation protocol was optimized for Thermus thermophilus HB27, which shows the highest natural competence efficiency within the genus and the highest DNA uptake rates. Other strains with less competence capability should be treated with larger quantities of DNA and/or longer incubation periods.
- In case transformation has been performed to introduce a mutation, transformants should be re-streaked on selective plates and further grown in liquid under each particular condition to ensure a stable mutation.
- DNA amount required varies from 10 ng when employing genomic DNA to 1,000 ng when employing PCR products. On average, replicative plasmids are employed at 100-250 ng.
- Regardless the selective marker employed, post-incubation times are at least 3 to 4 h at 65 °C and 150 rpm. If DNA is added at the start of the exponential phase (OD600 = 0.2), times delays up to 5 h incubation.
Recipes
- TB liquid medium
0.8 % tryptone (w/v)
0.4 % yeast extract (w/v)
50 mM NaCl, pH 7.5
Thermus water (carbonated-rich mineral water)*
pH may be adjusted by the addition of NaOH - TB agar plates
0.8% tryptone (w/v)
0.4% yeast extract (w/v)
50 mM NaCl, pH 7.5
1.8% agar (grade A) (w/v)
Thermus water (carbonated-rich mineral water)*
*Note: The analytical composition of the spring mineral water employed, per mg/ml, is listed beneath (Table 1). Other mineral waters easily purchasable such as Evian water or a laboratory solution resembling this recipe, can be employed.
Table 1. Mineral composition of the spring water from Jaraba Spring in Zaragoza (Spain)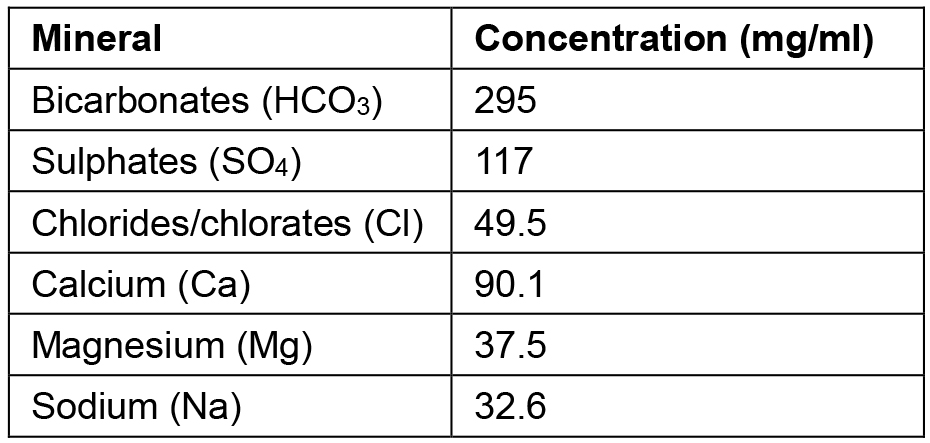
Acknowledgments
This work has been funded by Grants BIO2013-44963-R, and FP7-PEOPLE-2012-IAPP No. 324439. A. Blesa held an FPI contract from the Spanish Ministry of Economy and Competitivity.
References
- Averhoff, B. (2009). Shuffling genes around in hot environments: the unique DNA transporter of Thermus thermophilus. FEMS Microbiol Rev 33(3): 611-626.
- Brouns, S. J., Wu, H., Akerboom, J., Turnbull, A. P., de Vos, W. M. and van der Oost, J. (2005). Engineering a selectable marker for hyperthermophiles. J Biol Chem 280(12): 11422-11431.
- César, C. E., Alvarez, L., Bricio, C., van Heerden, E., Littauer, D. and Berenguer, J. (2011). Unconventional lateral gene transfer in extreme thermophilic bacteria. Int Microbiol 14(4): 187-199.
- Gold, V. A., Salzer, R.; Averhoff, B. and Kühlbrandt, W. (2015). Structure of a type IV pilus machinery in the open and closed state. Elife 4.
- Koyama, Y., Hoshino, T., Tomizuka, N. and Furukawa, K. (1986). Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J Bacteriol 166(1): 338-340.
- Lasa, I., Caston, J. R., Fernandez-Herrero, L. A., de Pedro, M. A. and Berenguer, J. (1992). Insertional mutagenesis in the extreme thermophilic eubacteria Thermus thermophilus HB8. Mol Microbiol 6(11): 1555-1564.
- Nakamura, A., Takakura, Y., Kobayashi, H. and Hoshino, T. (2005). In vivo directed evolution for thermostabilization of Escherichia coli hygromycin B phosphotransferase and the use of the gene as a selection marker in the host-vector system of Thermus thermophilus. J Biosci Bioeng 100(2): 158-163.
- Schwarzenlander, C. and Averhoff, B. (2006). Characterization of DNA transport in the thermophilic bacterium Thermus thermophilus HB27. FEBS J 273(18): 4210-4218.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Blesa, A. and Berenguer, J. (2016). Transformation of Thermus Species by Natural Competence. Bio-protocol 6(22): e2007. DOI: 10.21769/BioProtoc.2007.
Category
Microbiology > Microbial genetics > Transformation
Molecular Biology > DNA > Transformation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








