- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantitative Measurements of HIV-1 and Dextran Capture by Human Monocyte-derived Dendritic Cells (MDDCs)
Published: Vol 6, Iss 22, Nov 20, 2016 DOI: 10.21769/BioProtoc.2004 Views: 11068
Reviewed by: Kristopher MarjonVaibhav B ShahValeria Lulla

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Fluorescence Lifetime-Based Separation of FAST-Labeled Cellular Compartment
Aidar R. Gilvanov [...] Yulia A. Bogdanova
Oct 5, 2025 1325 Views

Protocol for the Isolation and Analysis of Extracellular Vesicles From Peripheral Blood: Red Cell, Endothelial, and Platelet-Derived Extracellular Vesicles
Bhawani Yasassri Alvitigala [...] Lallindra Viranjan Gooneratne
Nov 5, 2025 1449 Views

An Ex Vivo Lung Histoculture Model for Studying Pulmonary Infection and Immune Response With SARS-CoV-2 as an Example of RNA Virus
Elena V. Maryukhnich [...] Elena J. Vasilieva
Dec 20, 2025 751 Views
Abstract
The aim of this protocol is to describe how to measure and quantify the amount of HIV-1 particles and dextran molecules internalized in human monocyte derived dendritic cells (MDDCs), using three different techniques: flow cytometry, quantitative PCR and confocal microscopy.
Background
This protocol was developed in order to assess the changes of HIV-1 internalization upon disruption of actin nucleation in human monocyte derived dendritic cells. Following a shRNA screen to identify genes important for HIV-1 transfer from dendritic cells to T cells, we observed that a disruption of actin nucleation leads to a switch from actin rich dendrites to blebs, due to an excess of actomyosin contraction. As a consequence, a decrease of HIV-1 transfer and an increase of HIV-1 internalization due to bleb retraction-driven macropinocytosis were observed. We concluded that effectors of actin nucleation and stabilization were key to maintain HIV-1 on actin-rich dendrites and to limit its endocytosis, for efficient transfer to T lymphocytes (Menager and Littman, 2016).
Materials and Reagents
- Production of HIV-1 particles
- 10 cm plate (100 x 20 mm) (Corning, Falcon®, catalog number: 353003 )
- 15 cm plate (150 x 20 mm) (Corning, Falcon®, catalog number: 353025 )
- 15 ml conical tube
- 33 mm Millex-HV syringe filter, PVDF, 0.45 μm (EMD Millipore, catalog number: SLHV033RS )
- 60 ml syringe with BD Luer-LokTM tip (BD, Luer-LokTM, catalog number: 309653 )
- 50 ml high clarity PP centrifuge tube, conical bottom, sterile, 25/bag, 500/case (Corning, Falcon®, catalog number: 352070 )
- 293FT cells (Thermo Fisher Scientific, InvitrogenTM, catalog number: R700-07 )
- X4-HIV-Gag-iGFP plasmid
Note: Derived from HIV-1 molecular clone NL4-3 modified to insert the GFP protein in between the MA and CA domains of Gag as previously described (Hubner et al., 2007). - Dulbecco’s modification of Eagles medium (DMEM) (Mediatech, catalog number: 10-017-CV )
- Fetal bovine serum (FBS)
- MEM amino acid solution (GE Healthcare, HyCloneTM, catalog number: SH30598.01 )
- HyCloneTM L-glutamine (GE Healthcare, HyCloneTM, catalog number: SH3003401 )
- HyCloneTM penicillin streptomycin 100x solution (GE Healthcare, HyCloneTM, catalog number: SV30010 )
- Gentamicin (50 mg/ml) (Thermo Fisher Scientific, GibcoTM, catalog number: 15750060 )
- Geneticin® selective antibiotic (G418 sulfate) (powder) (Thermo Fisher Scientific, GibcoTM, catalog number: 11811023 )
- Trypsin 0.05% protease solution with porcine trypsin, HBSS, EDTA; without calcium, magnesium (GE Healthcare, HyCloneTM, catalog number: SH30236.01 )
- ProFection® mammalian transfection system-calcium phosphate (Promega, catalog number: E1200 )
- Calcium chloride (CaCl2)
- 100 mM sodium pyruvate solution (GE Healthcare, HyCloneTM, catalog number: SH30239.01 )
- 1 M HEPES solution (GE Healthcare, HyCloneTM, catalog number: SH30237.01 )
- D10 medium (see Recipes)
- 10 cm plate (100 x 20 mm) (Corning, Falcon®, catalog number: 353003 )
- Differentiation of monocyte derived dendritic cells (MDDCs)
- 60 ml syringe with BD Luer-LokTM tip (BD, Luer-LokTM, catalog number: 309653 )
- Needle (50/sp, 500/ca), 18 G x 1 ½ in. (BD, SafetyGlideTM, catalog number: 305918 )
- 3 ml plastic disposable transfer (Pasteur) pipettes, sterile in 20s (Elkay Laboratory Products, Liquipette®, catalog number: 127-P503-20S )
- 40 μm nylon cell strainer
- LS columns (Miltenyi Biotec, catalog number: 130-042-401 )
- 10 cm plate (100 x 20 mm) (Corning, Falcon®, catalog number: 353003 )
- 50 ml buffy coat from New York Blood Center (leukapharesis)
- Ficoll-Paque PLUS (GE Healthcare, HyCloneTM, catalog number: 17-1440-02 )
- Dulbecco’s phosphate buffered saline (DPBS), without calcium, magnesium, phenol red (GE Healthcare, HyCloneTM, catalog number: SH30028.02 )
- Bovine serum albumin (BSA), fraction V, heat shock (Roche Diagnostics, catalog number: 03116956001 )
- EDTA
- RPMI 1640 media (GE Healthcare, HyCloneTM, catalog number: SH30096.01 )
- Fetal bovine serum (FBS)
- 1 M HEPES solution (GE Healthcare, HyCloneTM, catalog number: SH30237.01 )
- 2-mercaptoethanol (55 mM) (1,000x) (Thermo Fisher Scientific, GibcoTM, catalog number: 21985-023 )
- HyCloneTM L-glutamine (GE Healthcare, HyCloneTM, catalog number: SH3003401 )
- HyCloneTM penicillin streptomycin 100x solution (GE Healthcare, HyCloneTM, catalog number: SV30010 )
- Geneticin® selective antibiotic (G418 sulfate) (powder) (Thermo Fisher Scientific, GibcoTM, catalog number: 11811023)
- Anti-human CD14 microbeads
- 20.Human GM-CSF (Affymetrix, eBioscience, catalog number: 34-8339-82 )
- Human IL-4 (Affymetrix, eBioscience, catalog number: 34-8049-82 )
- Anti-hDC-SIGN PE (Clone: 120507), mouse IgG2B (R&D Systems, catalog number: FAB161P )
- Anti-hCD14 APC (Clone: 61D3) (Affymetrix, eBioscience, catalog number: 17-0149-42 )
- MACS buffer (see Recipes)
- DC medium (see Recipes)
- 60 ml syringe with BD Luer-LokTM tip (BD, Luer-LokTM, catalog number: 309653 )
- MDDC loading with HIV-1 or dextran
- 96 well clear round bottom TC-treated cell culture microplate, with lid, individually wrapped, sterile (Corning, Falcon®, catalog number: 353077 )
- X4-HIV-Gag-iGFP plasmid
Note: Derived from HIV-1 molecular clone NL4-3 modified to insert the GFP protein in between the MA and CA domains of Gag as previously described (Hubner et al., 2007) - Dextran, fluorescein (10,000 MW, anionic, lysine fixable) (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: D-1820 )
- Dextran, fluorescein (70,000 MW, anionic, lysine fixable) (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: D-1822 )
- Dextran, fluorescein (500,000 MW, anionic, lysine fixable) (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: D-7136 )
- Dextran, fluorescein (2,000,000 MW, anionic, lysine fixable) (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: D-7137 )
- Dulbecco’s phosphate buffered saline (DPBS), without calcium, magnesium, phenol red (GE Healthcare, HyCloneTM, catalog number: SH30028.02 )
- MACS buffer (see Recipes)
- 96 well clear round bottom TC-treated cell culture microplate, with lid, individually wrapped, sterile (Corning, Falcon®, catalog number: 353077 )
- Analysis of HIV-1 or dextran capture by flow cytometry
- Hanks’ balanced salt solution (HBSS) (1x) (Mediatech, catalog number: 21-023-CV )
- LIVE/DEAD® Fixable Blue Dead Cell Stain Kit (for UV excitation) (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: L23105 )
- Anti-hDC-SIGN PE (Clone: 120507), mouse IgG2B (R&D Systems, catalog number: FAB161P )
- Anti-hCD14 APC (Clone: 61D3) (Affymetrix, eBioscience, catalog number: 17-0149-42 )
- Fixation/Permeabilization Solution Kit (RUO) (Fixation/Permeabilization solution 125 ml and BD Perm/WashTM buffer 100 ml) (BD, Cytofix/CytopermTM, catalog number: 554714 )
- Anti-P24 KC57-RD1 (Beckman Coulter, catalog number: 6604667 )
- Hanks’ balanced salt solution (HBSS) (1x) (Mediatech, catalog number: 21-023-CV )
- Analysis of HIV-1 capture by Quantitative PCR (QPCR)
- LightCycler® 480 multiwell plate 96, white (Roche Diagnostics, catalog number: 04729692001 )
- LightCycler® 480 SYBR Green I Master (Roche Diagnostics, catalog number: 04887352001 )
- TRIzol® reagent (Thermo Fisher Scientific, AmbionTM, catalog number: 15596-018 )
- SuperScript® III first-strand synthesis system (Thermo Fisher Scientific, InvitrogenTM, catalog number: 18080-051 )
- primers:
GFP forward: 5’-ACGTAAACGGCCACAAGTTC-3’
GFP Reverse: 5’-AAGTCGTGCTGCTTCATGTG-3’
GAPDH forward: 5’-CCCATCACCATCTTCCAGGAGCG-3’
GAPDH Reverse: 5’-GAGATGATGACCCTTTTGGCTCC-3’
- LightCycler® 480 multiwell plate 96, white (Roche Diagnostics, catalog number: 04729692001 )
- Analysis of HIV-1 or dextran capture by confocal microscopy
- FisherbrandTM microscope slides (25 x 75 x 1.0 mm, double frosted precleaned) (Thermo Fisher Scientific, Fisher Scientific, catalog number: 12-552-5 )
- Cover slip, round, 5 mm diameter, #1 0211 glass (Corning, Falcon®, catalog number: CLS-1763-005 )
- Poly-L-lysine solution (Sigma-Aldrich, catalog number: P8920-100ML )
- Paraformaldehyde, 16% solution, EM grade (Electron Microscopy Sciences, catalog number: 15710 )
- PIPES (1,4-piperazinediethanesulfonic acid) (Sigma-Aldrich, catalog number: P6757-100G )
- EGTA [Ethylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid tetrasodium salt, ≥ 97%] (Sigma-Aldrich, catalog number: E8145-10G )
- Magnesium sulfate (MgSO4)
- Potassium hydroxide (KOH)
- TritonTM X-100 (Sigma-Aldrich, catalog number: X100-100ML )
- 5% casein solution as described and prepared in Dustin et al. (2007)
- 4’,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: D1306 )
- 1,4-diazabicyclo[2.2.2]octane (DABCO) (Sigma-Aldrich, catalog number: D27802-25G )
- Poly(vinyl alcohol) (PVA) (Sigma-Aldrich, catalog number: 341584-25G )
- Glycerol (Sigma-Aldrich, catalog number: G5516-100ML )
- Tris buffer, pH 8.7 (1.5 M) (Bio-Rad Laboratories, catalog number: 1610798 )
- 2x PHEM buffer (see Recipes)
- DABCO-PVA (see Recipes)
- FisherbrandTM microscope slides (25 x 75 x 1.0 mm, double frosted precleaned) (Thermo Fisher Scientific, Fisher Scientific, catalog number: 12-552-5 )
Equipment
- Centrifuge (Eppendorf, model: 5810 )
- Vortex
- CO2 incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: HeracellTM 240 )
- Flow cytometer (BD, model: LSRII )
- High throughput sampler (HTS) (BD)
- QPCR system (Roche Diagnostics, model: LightCycler 480 )
- Confocal microscope (Zeiss, model: LSM 710 )
Software
- ImageJ software
Procedure
- Production of HIV-1
Note: HIV-1 particles are produced after calcium phosphate transfection in 293FT cells.- 293FT cell culture
- 293FT cells are cultured in D10 medium in 10 cm plates, in the presence of 500 μg/ml of Geneticin®.
- When approaching confluence, cells are trypsinized (0.05% trypsin), washed by centrifugation (475 x g, 5 min at 4 °C) and plated after 1/5 dilution in 10 cm plates.
- Plate 8 x 106/15 cm plate, 293FT cells the day before transfection in 30 ml of D10 medium without Geneticin®.
- One 15 cm plate is going to produce 20 ml of virus, so transfection experiment should be scaled accordingly.
- 293FT cells are cultured in D10 medium in 10 cm plates, in the presence of 500 μg/ml of Geneticin®.
- Calcium phosphate transfection
Note: The Promega kit is used for calcium phosphate transfection. Transfection protocol is described for transfection in one 15 cm plate.- Dilute 60 μg of DNA (X4-HIV-Gag-iGFP) with 1.12 ml nuclease-free H2O in a 15 ml conical tube (tube 1).
- Add 168 μl of CaCl2, 2 M, to the diluted DNA.
- In another tube (tube 2), add 1,350 μl of 2x HEPES-buffered saline (2x HBS).
- While gently vortexing tube 2, add dropwise the prepared DNA solution to the HBS in tube 2.
- Incubate the solution at room temperature for 30 min.
- Vortex the transfection solution again just before adding it to the cells.
- Add the solution containing DNA- CaCl2-HBS, dropwise in various places around the plate.
- Swirl the plate to distribute the solution evenly.
- 20 h after transfection, carefully replace the culture medium with 20 ml of D10 medium without G418.
- 24 h after medium change, harvest the supernatant.
- Filter the supernatant twice (to remove cells and debris) with 0.45 μm sterile filter unit with PVDF membrane.
- Filtered viral supernatant can be used fresh or stored at -80 °C.
- Dilute 60 μg of DNA (X4-HIV-Gag-iGFP) with 1.12 ml nuclease-free H2O in a 15 ml conical tube (tube 1).
- 293FT cell culture
- Differentiation of monocyte derived dendritic cells (MDDCs)
- Obtaining peripheral blood mononuclear cells (PBMCs). PBMCs are obtained from buffy coats ordered from the New York Blood Center using mononuclear cell isolation with Ficoll-Paque separation media.
- Ficoll and DPBS are brought to room temperature (RT).
- Recover blood from buffy coat using 60 ml syringe and 18 G thick needle.
- Mix blood with RT DPBS (1:1 ratio).
- Add 25 ml of diluted blood using a needle to a 50 ml Falcon tube already containing 15 ml of Ficoll.
- Centrifuge for 30 min at 845 x g, RT and with brake off.
- Recover the white leukocyte layer (in between the yellow serum and the red platelets layers) with a plastic pipette.
- Wash the leukocyte layers containing the PBMCs with cold DPBS, centrifuge at 170 x g for 10 min at 4 °C (slow spin to remove platelets).
- Resuspend the cells in MACS buffer, wash, spin at 475 x g, 5 min at 4 °C.
- Count the cells.
- Add 50 μl of anti-human CD14 microbeads from Miltenyi and 950 μl MACS buffer per 100 million PBMCs.
- 20 min incubation at 4 °C.
Note: While doing the staining of PBMCs with anti CD14 magnetic beads, always do the staining in the fridge rather than on ice in order to prevent antibody ‘capping effect’.
- Wash in MACS buffer, then spin at 475 x g, 5 min at 4 °C.
- Resuspend in 5 ml MACS buffer and filter using a 40 μm nylon cell strainer.
- Run the sample through an LS MACS separation column for positive selection.
- Perform 3 washes of the LS column, using 3 ml of MACS buffer.
- Recover the CD14+ cells from the column by using 5 ml of MACS buffer and a plunger.
- Count the cells and plate them at a concentration of 1 million/ml in DC medium with hGM-CSF (10 ng/ml) and hIL-4 (50 ng/ml).
Note: To obtain optimal results regarding monocyte differentiation into MDDCs, plate 20 million monocytes per 10 cm plate, in 20 ml DC medium. - 2 days post isolation, add 40% of new DC medium with hGM-CSF (10 ng/ml) and hIL-4 (50 ng/ml).
- 4 days post isolation, check MDDC differentiation by flow cytometry: monitor upregulation of DC-SIGN and downregulation of CD14 (1/200 of stock solution for both antibodies).
- Obtaining peripheral blood mononuclear cells (PBMCs). PBMCs are obtained from buffy coats ordered from the New York Blood Center using mononuclear cell isolation with Ficoll-Paque separation media.
- MDDC loading with HIV-1 or dextran
- Plate 50,000 MDDCs (1 million/ml) per well in a U bottom 96-well plate.
- Place the 96-well plate on ice, while adding HIV-1 or dextran molecules, for synchronization of capture.
- Add 50 ng of HIV-1 or 100 μg/ml of FITC-labeled dextran molecules of various size (10kDa, 70kDa, 500kDa, 2,000kDa).
Note: Don’t forget to add negative controls: no virus or dextran and/or cells staying at 4 °C. - Incubate at 37 °C, 5% CO2, for 4 and 24 h for HIV-1 and 30 min and 4 h for dextran molecules.
- After incubation, wash extensively by adding 200 μl cold DPBS and centrifuge 5 min, 475 x g at 4 °C.
- Repeat the washing step 3 times.
- Analyze HIV-1 or dextran capture.
- Plate 50,000 MDDCs (1 million/ml) per well in a U bottom 96-well plate.
- Analysis of HIV-1 or dextran capture by flow cytometry
- For flow cytometry analysis, take the cells loaded with dextran or HIV-1.
- Stain with 50 μl of a live/dead blue fixable dye diluted in HBSS (1x) (1/500) 15 min on ice.
- Wash using MACS buffer, 475 x g, 5 min, 4 °C.
- Stain with anti-hDC-SIGN-PE (1/200 diluted in MACS buffer) and anti-hCD14-APC (1/200 diluted in MACS buffer) to control for MDDC differentiation.
- Incubate for 30 min, 4 °C, covered.
- Wash using MACS buffer, 475 x g, 5 min, 4 °C.
- Resuspend in 100 μl.
- For dextran detection, no further staining is required. For HIV-1 detection, cells need to be fixed, using 50 μl of BD Cytofix/CytopermTM, 20 min at 4 °C.
- Wash using 100 μl of BD perm/Wash buffer.
- Stain for HIV-1 capsid using anti-P24-RD1, diluted 1/500 in BD perm/Wash buffer. 1 h at RT.
- Wash using MACS buffer, 475 x g, 5 min, 4 °C.
- Resuspend in 100 μl.
- Proceed to flow cytometry analysis.
Note: For flow cytometry, BD high throughput sampler (HTS) can be used to run 96-well plates.
- For flow cytometry analysis, take the cells loaded with dextran or HIV-1.
- Analysis of HIV-1 capture by Quantitative PCR (QPCR)
Real time quantitative PCR (qPCR) was performed after reverse transcription using a Roche LightCycler 480 with Roche 480 SYBR Green I Master reagent according to manufacturer specifications. The relative abundance of HIV is calculated based on the amount of GFP mRNA (the virus encodes GFP) using a standard curve and normalized using GAPDH as a control.- Total RNA is extracted from 3 x 50,000 MDDCs using TRIZOL® RNA isolation protocol.
- 5 μl of RNA is reverse transcribed using SuperScript® III first-strand synthesis system in a 20 μl final reaction volume.
- For each sample, 2 μl of diluted cDNA (dilution in water, 1/2 and 1/5) is used for QPCR reaction.
- QPCR mix:
6 μl of 2x SYBR Green mix
0.6 μl primer F (10 μM)
0.6 μl primer R (10 μM)
2.8 μl of H2O
2 μl of cDNA - QPCR reaction:
Step 1: 95 °C, 5 min
Step 2: 95 °C, 5 sec
Step 3: 60 °C, 30 sec
Step 4: 72 °C, 20 sec
Back to step 2, 50 cycles - QPCR analysis: based on standard curve, determine GFP mRNA quantity and normalize it by GAPDH in each sample.
- Total RNA is extracted from 3 x 50,000 MDDCs using TRIZOL® RNA isolation protocol.
- Analysis of HIV-1 or dextran capture by confocal microscopy (Figures 1 and 2)
- MDDCs loaded with HIV-1 or dextran are put on coverslips previously coated with poly-L-lysine solution, for 30 min at 37 °C.
- Phalloidin staining (to detect cortical actin), or any cell surface staining, is performed in PBS-1% BSA for 30 min at room temperature (Figure 2).
- For fixation, incubate with warm paraformaldehyde A (PFA) (2%) in PHEM buffer, for 10 min at room temperature.
- Permeabilize in 0.1% Triton X-100 diluted in PHEM buffer, for 5 min at RT.
- Blocking is done in 5% casein solution, 1 h at 4 °C.
- For intracellular staining (if needed), antibodies are diluted in 5% casein solution and incubated for 1 h at room temperature.
- For nuclear detection, cells are then stained with DAPI, (1/5,000 dilution from stock at 5 mg/ml in H2O), at RT (Figures 1 and 2).
- Upon 5 washing (200 μl each) in PBS-0.1% BSA, cells are mounted in a homemade DABCO-PVA medium.
- Using a confocal microscope, analyze for each cell the intensity of internalized GFP or FITC, which represent, respectively, captured HIV-1 particles and dextran molecules. To do so, acquire 400 nm Z-stacks and determine the HIV-1 and dextran molecules. Quantification of HIV-1 and dextran uptake will be discussed in the Data analysis section. (Figures 1 and 2)
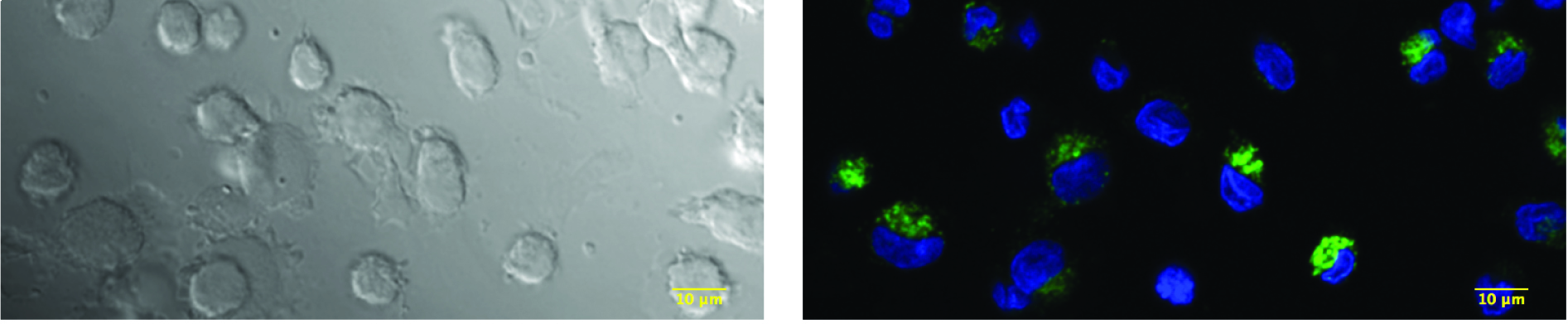
Figure 1. Example of untransduced and untreated MDDCs loaded for 4 h with 100 μg/ml of 10 kDa FITC-labeled dextran molecules. Cells on coverslips can be seen on the left panel (bright-field). On the right panel, one Z-stack section of 400 nm is displayed where nuclei of cells were stained with DAPI (blue) and molecules of dextran can be visualized in green.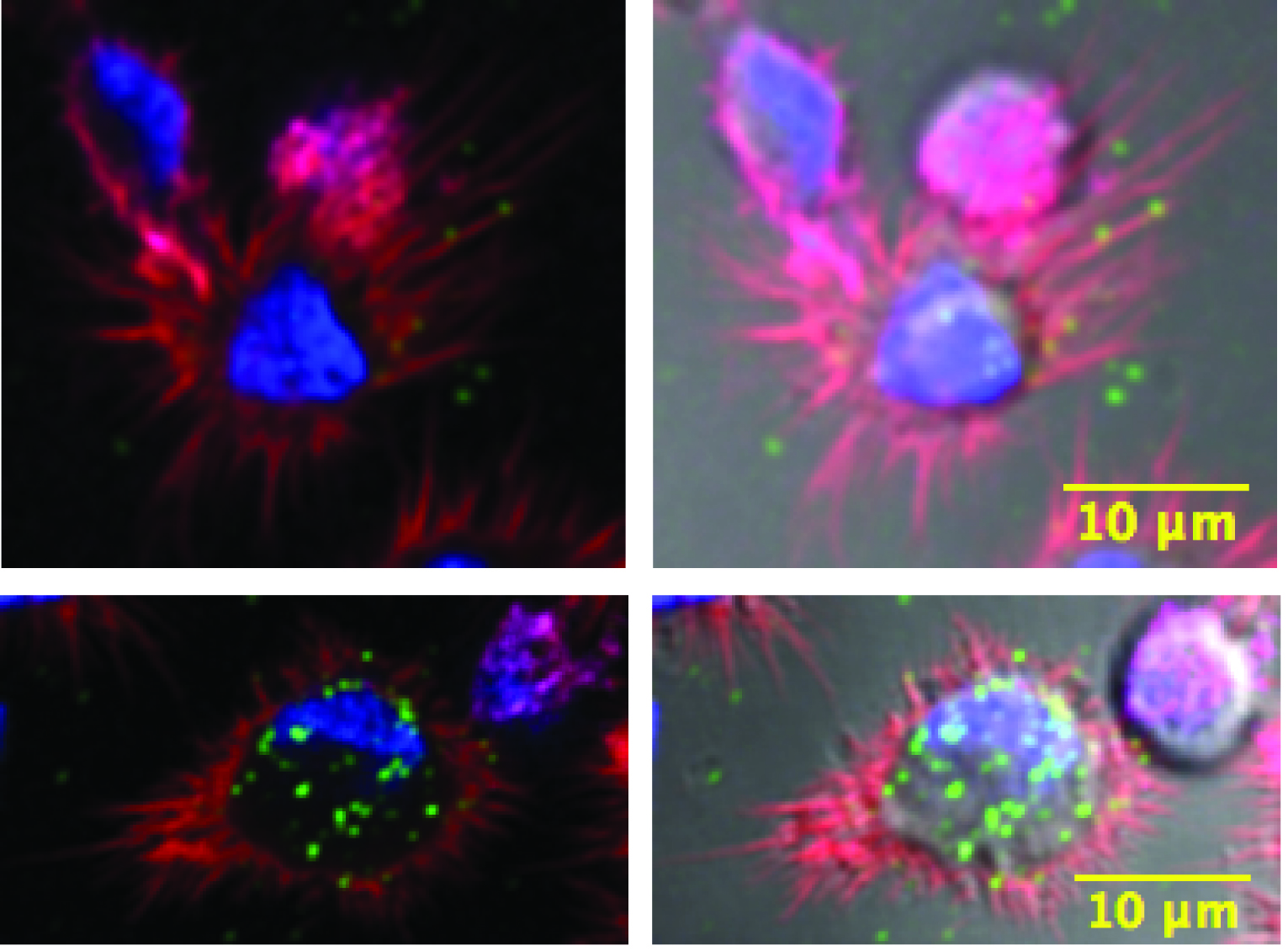
Figure 2. Example of HIV-1 internalization by confocal microscopy. MDDCs untransduced (top panel) or transduced with a shRNA against DNM2 (bottom panel) loaded for 4 h with 50 ng of HIV-1 (green dots). Bright-field can be seen on the left panel. On the right panel, one Z-stack section of 400 nm is displayed where nuclei of cells were stained with DAPI (blue), actin filaments in red, after phalloidin staining and captured HIV-1 in green.
- MDDCs loaded with HIV-1 or dextran are put on coverslips previously coated with poly-L-lysine solution, for 30 min at 37 °C.
Data analysis
- Analysis of HIV-1 or dextran capture by flow cytometry (Figure 3)
- First, the gate must be set on live cells, by excluding cells that have incorporated the live/dead stain. It is important that MDDCs are well differentiated, as assessed by high expression of DC-SIGN and low expression of CD14. (Figure 3A)
- Monocytes are usually CD14 high and DC-SIGN low.
- The amount of HIV-1 or dextran internalized can be calculated by comparing the mean fluorescence intensity (MFI) to the negative control and determining the ratio of internalization between samples (Figure 3B).
- It is recommended that HIV and dextran capture be assessed by using different dilutions and different times of incubation, in order to draw a representative and more accurate curve.
- Each sample is usually analyzed in triplicate and the experiment is performed with cells coming from at least 3 independent healthy blood donors.
- Statistical analyses of the differences in MFI between samples can be done using the Holm-Sidak multiple comparison test following one-way ANOVA (NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
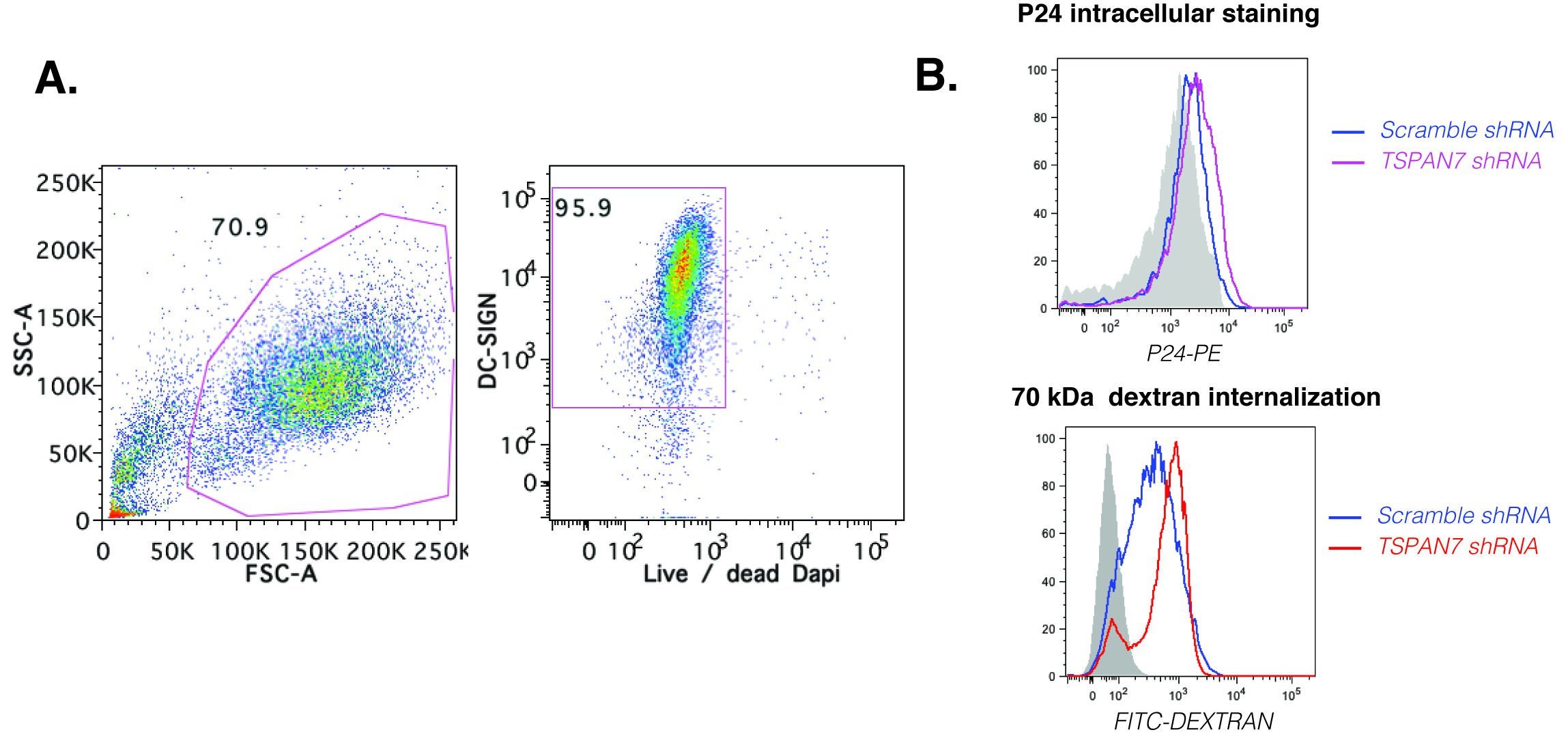
Figure 3. Analysis of HIV-1 and dextran capture by flow cytometry. A. Gating strategy to analyse MDDCs: left panel, gating on MDDCs based on size (FSC) and granularity (SSC); right panel gating on alive differentiated MDDCs (DC-SIGN+; dapi-). B. Example of histograms generated after gating on MDDCs loaded with HIV-1 (top panel) or dextran (lower panel). In grey, an isotype control for P24 is used to assess the amount of HIV-1 captured by MDDCs transduced with scramble shRNA vs. TSPAN7 shRNA. For dextran internalization, the grey histograms represent a negative control where cells are staying at 4 °C to prevent internalization.
- First, the gate must be set on live cells, by excluding cells that have incorporated the live/dead stain. It is important that MDDCs are well differentiated, as assessed by high expression of DC-SIGN and low expression of CD14. (Figure 3A)
- Analysis of HIV-1 capture by Quantitative PCR (QPCR) (Figure 4)
- After reverse transcription, the amount of HIV-1 RNA was detected by quantitative PCR with primers for GFP (encoded inside the HIV genome) and normalized to GAPDH.
- To assess the quantity in the most accurate way possible, every experiment should include a standard curve using the GFP primers and also the GAPDH primers.
- Then the standard curve is used to determine the quantity of HIV internalized in each sample and compare that to the negative controls (no virus or capture experiment performed at 4 °C).
- Normalized HIV-1 quantity can be compared between different samples to assess the increase or the decrease in HIV capture (Figure 4).
- Due to experimental variations that could exist between different blood donors, it is preferable not to compare samples coming from different donors, but seek consistency between donors in terms of trends. Experiments should be done in triplicate and in at least five different blood donors.
- Statistical analyses of the differences in HIV capture between samples can be done using the Holm-Sidak multiple comparison test following one-way ANOVA (NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001) (Figure 4).
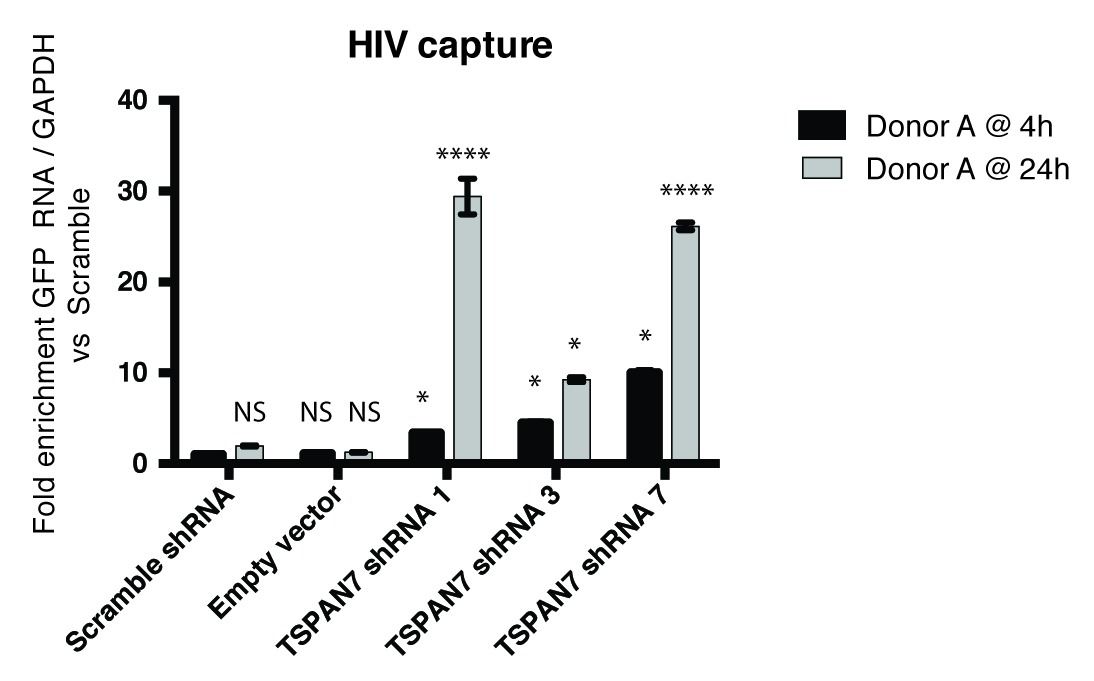
Figure 4. Analysis of HIV-1 capture by quantitative PCR. Example of histograms after HIV capture experiments and analysis by quantitative PCR. MDDCs transduced with scramble shRNA, empty vectors or shRNA against TSPAN7 were loaded with HIV-GFP for 4 or 24 h. Quantity of HIV-1 captured is analysed by PCR against GFP, and compared to the quantity captured after scramble shRNA (set arbitrarily to a value of 1).
- After reverse transcription, the amount of HIV-1 RNA was detected by quantitative PCR with primers for GFP (encoded inside the HIV genome) and normalized to GAPDH.
- Analysis of HIV-1 or dextran capture by confocal microscopy (Figure 5)
- For quantification of dextran internalization by confocal microscopy, we use phalloidin staining (for filamentous actin) to identify the outside borders of every cell.
- Using the ImageJ software, the maximal intensity of dextran per pixel is collected for each Z-stack.
- The total intensity of dextran is then obtained and normalized to the volume analyzed.
- The integrated intensity can be calculated by taking the sum of all voxels, at each intensity greater than background (intensity ≥ 200), then multiply by the intensity and summing.
- Volume is obtained by counting all voxels greater than background.
- Concentration is the integrated intensity divided by volume (arbitrary units) (Figure 5).
- For each sample, we recommend to calculate the concentration of dextran for at least 100 cells and to repeat the experiment with at least 3 different human blood donors. Unpaired t-test can be used to determine statistical significance.
- Using the ImageJ software, the maximal intensity of dextran per pixel is collected for each Z-stack.
- Regarding quantification of HIV-1 internalization, 400 nm Z-stacks are acquired using a confocal microscope with an average of 20 Z-stacks per cell.
- Every HIV-1 particle (green dot) can be manually counted as internalized based on its localization compared to the cortical actin barrier (phalloidin staining).
- Based on electron microscopy data, an average of 5 HIV viral particles was estimated to be present per HIV-1 aggregate.
- For each cell, the total of HIV-1 can thus be calculated and an average of at least 100 cells per sample is analyzed. Unpaired t-test can be used to determine statistical significance.
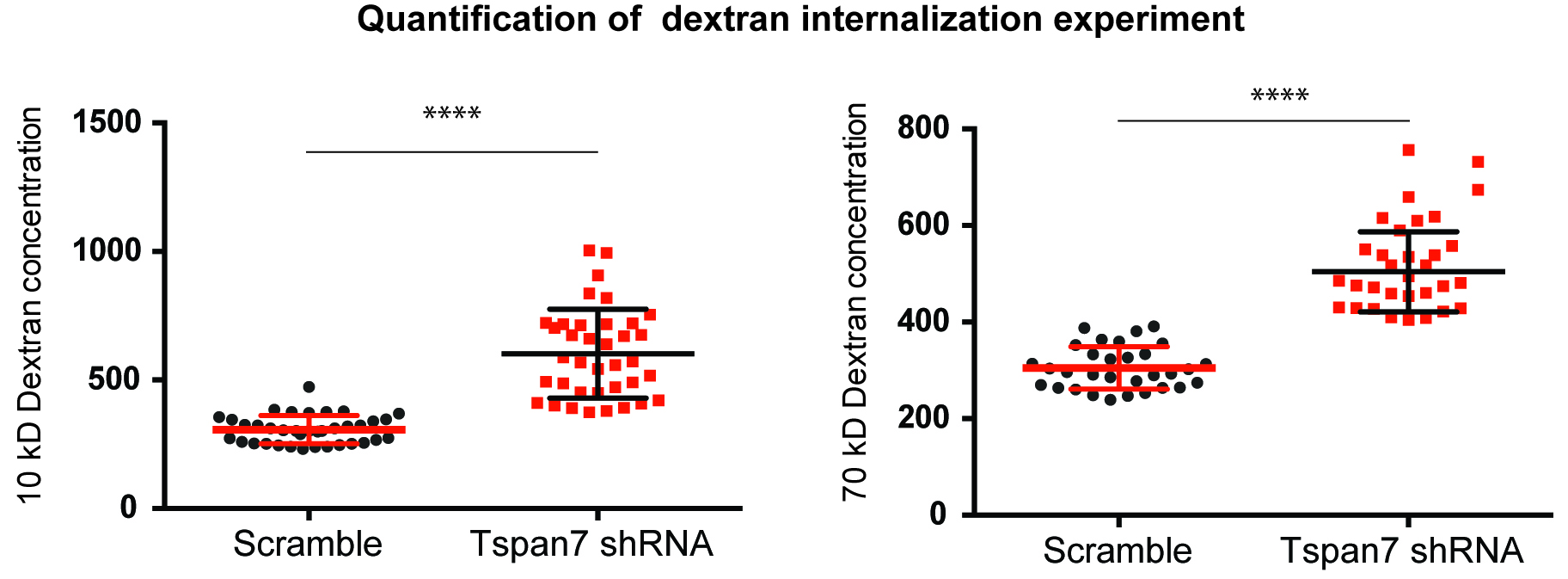
Figure 5. Analysis of dextran capture by confocal microscopy. MDDCs transduced with a scramble shRNA or shRNA against TSPAN7 were cultured for 4 h with 10 kDa (left panel) or 70 kDa molecules of dextran (right panel). The concentration of dextran internalized by each MDDC was then determined as explained above. Briefly, concentration is the integrated density of dextran staining divided by volume and is displayed with arbitrary units on the y axis. For each condition, each dot represents a different cell and the average is displayed ± SEM.
- Every HIV-1 particle (green dot) can be manually counted as internalized based on its localization compared to the cortical actin barrier (phalloidin staining).
- For quantification of dextran internalization by confocal microscopy, we use phalloidin staining (for filamentous actin) to identify the outside borders of every cell.
Notes
- In general, when working with human primary cells, you need to make sure to have internal controls in order to be able to normalize the data and to compare data coming from different blood donors. Variation can also be observed from donor to donor and increasing the number of different human blood donors usually helps to have better analysis of the data.
- We recommend to always check the status of maturation of MDDCs. Experiments as described in this protocol are performed using immature MDDCs. Once mature, the results obtained in terms of HIV-1 capture and dextran internalization are different. Maturation status can be determined by looking at CD86 and CD80 molecules by flow cytometry. Immature MDDCs should be CD80 and CD86 low.
- All 3 techniques measure HIV-1 internalization by MDDCs. Combining 3 different methods is increasing the chances to detect even small differences in capture and internalization and is increasing the robustness of the results. To detect changes in HIV-1 internalization, flow cytometry is maybe not the best method as it doesn’t seem to be as sensitive as QPCR or confocal microscopy to detect small amount of HIV internalization. Quantitative PCR is maybe the fastest and easiest method but is not allowing a direct comparison of HIV-1 and dextran internalization and any artefactual contamination could potentially biased the data. The most demanding technique is probably the confocal microscopy but allows the most rigorous comparison of both HIV-1 and dextran internalization between different samples.
Recipes
- D10 medium
DMEM
10% fetal bovine serum (FBS, heat inactivated)
0.1 mM MEM non-essential amino acids
6 mM L-glutamine
1 mM MEM sodium pyruvate
100 U/ml penicillin
100 μg/ml streptomycin
50 μg/ml gentamycin - MACS buffer
DPBS
0.5% bovine serum albumin (BSA)
1 mM EDTA - DC medium
RPMI
10% FBS (heat inactivated)
10 mM Hepes (Hyclone)
55 μM β-mercaptoethanol
6 mM L-glutamine
100 U/ml penicillin
100 μg/ml streptomycin
50 μg/ml gentamycin - 2x PHEM buffer (500 mls)
18.14 g PIPES
6.5 g HEPES
3.8 g EGTA
0.99 g MgSO4
Adjust pH to 7.0 with 10 N KOH - DABCO-PVA
2.5% Dabco
10% polyvinylalcohol (PVA)
5% glycerol
25 mM Tris buffer, pH 8.7
Acknowledgments
We thank Alice F. Liang, Michael Cammer and the NYULMC OCS Microscopy Core for the service provided for light microscopy; Wendy Lin for technical help; and Jarrod Johnson, Nicolas Manel, for their critical advice. This work was supported by fellowships from EMBO, the Cancer Research Institute, and the Philippe Foundation (M.M.M.); by the Howard Hughes Medical Institute (D.R.L.) and Helen and Martin Kimmel Center for Biology and Medicine (D.R.L.); and by grants from the National Institutes of Health (R21AI084633) (D.R.L.) and NCRR (S10RR023704-01A1).
References
- Dustin, M. L., Starr, T., Varma, R. and Thomas, V. K. (2007). Supported planar bilayers for study of the immunological synapse. Curr Protoc Immunol Chapter 18: Unit 18 13.
- Hubner, W., Chen, P., Del Portillo, A., Liu, Y., Gordon, R. E. and Chen, B. K. (2007). Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J Virol 81(22): 12596-12607.
- Menager, M. M. and Littman, D. R. (2016). Actin dynamics regulates dendritic cell-mediated transfer of HIV-1 to T cells. Cell 164(4): 695-709.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ménager, M. M. and Littman, D. R. (2016). Quantitative Measurements of HIV-1 and Dextran Capture by Human Monocyte-derived Dendritic Cells (MDDCs). Bio-protocol 6(22): e2004. DOI: 10.21769/BioProtoc.2004.
Category
Cell Biology > Cell imaging > Fluorescence
Immunology > Host defense > Human
Cell Biology > Cell-based analysis > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










