Microfluidics-free single-cell genomics with templated emulsification
Speaker: Iain Clark
Online live: Feb 06, 2024 12:00 PM EST Posted: Apr 2, 2024 Views: 8279
Abstract
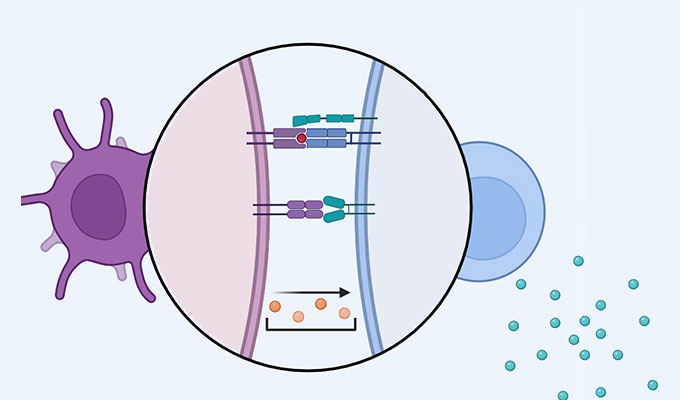
Current single-cell RNA-sequencing (scRNA-seq) methods utilize complex microfluidics or laborious multi-step fluid handling steps. To overcome these limitations, we developed a method called PIP-seq, which does not require specialized microfluidic devices, expertise, or hardware. Our approach uses particle-templated emulsification to generate water-in-oil droplets with a standard laboratory vortexer. This allows for the encapsulation of single cells with barcoded beads in less than two minutes. In previous work, we demonstrated that PIP-seq produces high-purity transcriptomes, is compatible with multi-omics measurements, and accurately reproduces scRNA-seq results from a commercial platform (10X Genomics). In this presentation, I will discuss the development of PIP-seq, its unique ability to accommodate a wide range of cell (10-106) and sample (1-103) numbers, and the new applications that this unlocks.
Highlights
• Developed a method to encapsulate single cells with barcode beads for scRNA-seq
• Uses a laboratory vortexer and no microfluidics
• Highly scalable (millions), fast (minutes), and user-friendly
• Commercialized by Fluent Biosciences (https://www.fluentbio.com/)
Speaker

Iain Clark, Ph.D.
Assistant Professor, Bioengineering, UC Berkeley
Iain C. Clark is an Assistant Professor in the Bioengineering Department at the University of California, Berkeley, and a ...
View more
References
Clark IC, Fontanez KM, Meltzer RH, Xue Y, Hayford C, May-Zhang A, D'Amato C, Osman A, Zhang JQ, Hettige P, Ishibashi JSA, Delley CL, Weisgerber DW, Replogle JM, Jost M, Phong KT, Kennedy VE, Peretz CAC, Kim EA, Song S, Karlon W, Weissman JS, Smith CC, Gartner ZJ, Abate AR. Microfluidics-free single-cell genomics with templated emulsification. Nat Biotechnol. 2023. Epub 20230306. doi: 10.1038/s41587-023-01685-z. PubMed PMID: 36879006.
Do you have a question about this webinar?
Post your question, and we'll invite the webinar speaker to respond. You're welcome to join the discussion by answering or commenting on questions ( Note: Not all questions, especially those not directly relevant to the webinar topic, may be answered by the speaker. ).
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question.
26 Q&A