- EN - English
- CN - 中文
Plasmodium berghei High-Throughput (PbHiT): a CRISPR-Cas9 System to Study Genes at Scale
(*contributed equally to this work, § Technical contact) 发布: 2026年01月20日第16卷第2期 DOI: 10.21769/BioProtoc.5572 浏览次数: 30
评审: Abhilash PadavannilAnonymous reviewer(s)
Abstract
Genetic modification is essential for understanding parasite biology, yet it remains challenging in Plasmodium. This is partially due to the parasite’s low genetic tractability and reliance on homologous recombination, since the parasites lack the canonical non-homologous end-joining pathway. Existing approaches, such as the PlasmoGEM project, enable genome-wide knockouts but remain limited in coverage and flexibility. Here, we present the Plasmodium berghei high-throughput (PbHiT) system, a scalable CRISPR-Cas9 protocol for efficient genome editing in rodent malaria parasites. The PbHiT method uses a single cloning step to generate vectors in which a guide RNA (gRNA) is physically linked to short (100 bp) homology arms, enabling precise integration at the target locus upon transfection. The gRNA also serves as a unique barcode, allowing pooled vector transfections and identification of mutants by downstream gRNA sequencing. The PbHiT system reliably recapitulates known mutant growth phenotypes and supports both knockout and tagging strategies. This protocol provides a reproducible and scalable tool for genome editing in P. berghei, enabling both targeted functional studies and high-throughput genetic screens. Additionally, we provide an online resource covering the entire P. berghei protein-coding genome and describe a step-by-step pooled ligation approach for large-scale vector production.
Key features
• PbHiT provides a high-throughput CRISPR-Cas9 genome editing platform optimised for Plasmodium berghei experimental infections in rodents.
• This protocol enables efficient and reproducible generation of knockout and tagged parasite lines using short homology arms.
• This protocol provides a free online resource for P. berghei gene construct design and requires basic knowledge of cloning.
• Transfection of Plasmodium berghei requires experience in handling mice/rats, an ethical permit, and an animal facility.
Keywords: Plasmodium bergheiGraphical overview
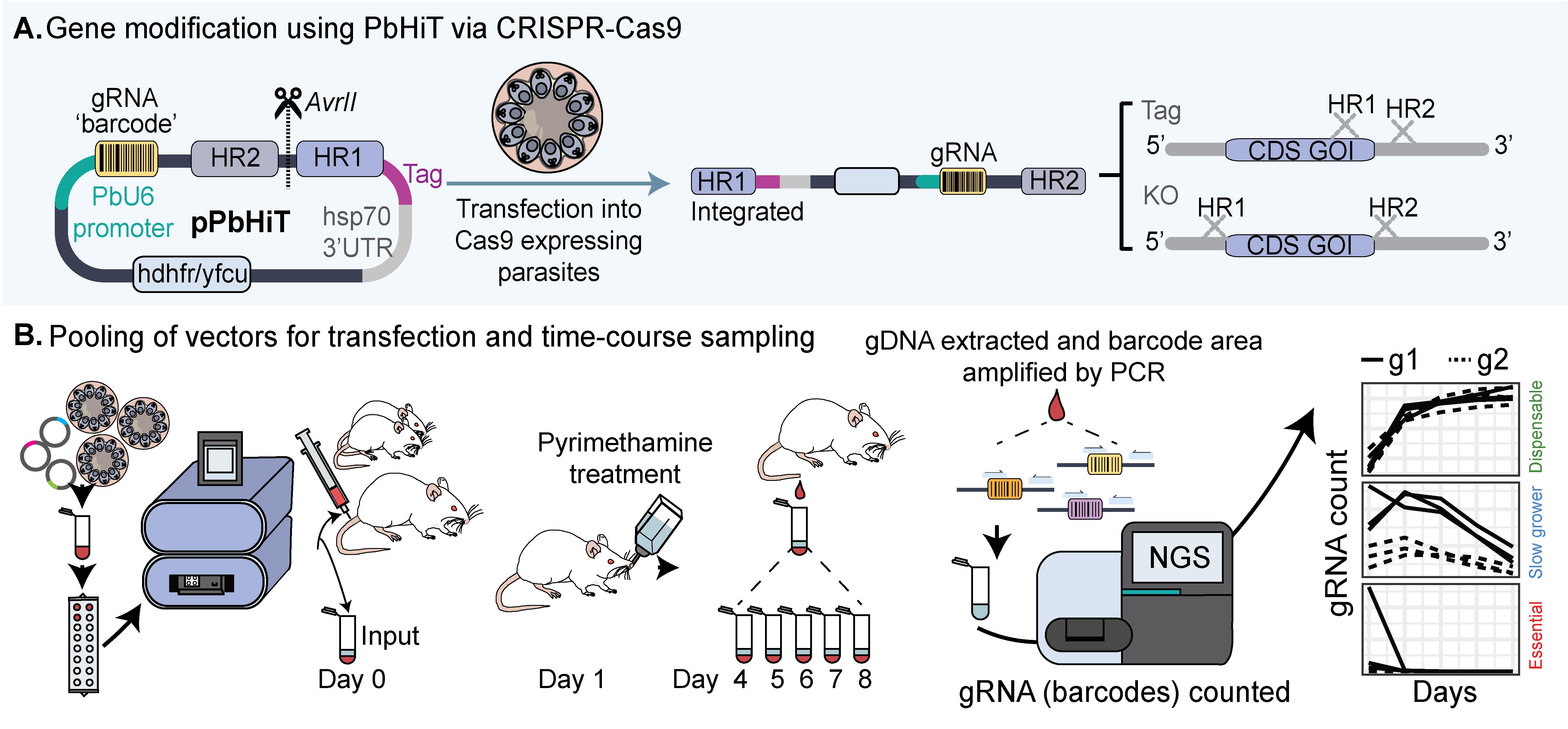
Schematic overview of the Plasmodium berghei high-throughput (PbHiT) workflow. (A) The pPbHiT vector (where p denotes plasmid) is designed with an hdhfr/yfcu drug selection cassette, facilitating both positive (hdhfr) and negative (yfcu) selection of the vector using pyrimethamine and 5-fluorocytosine, respectively. The synthetic fragment contains the gene-specific guide, homology region 2 (HR2) in the 3′ UTR of the target gene, and homology region 1 (HR1) either 100 bp upstream of the stop codon (tag) or start codon (knockout). The final vector is then linearised between HR2 and HR1 and integrated into the gene of interest in frame with the selected tag (Myc here), replacing the endogenous 3′ UTR with hsp70 3′UTR upon integration. (B) For scaling up, vectors are pooled together prior to transfection and injection into three individual mice. An input sample is taken on day 0, and then samples are taken daily once parasites are visible by blood smear (day 4 here), for 5 days. Genomic DNA (gDNA) is then extracted, and the barcode area is amplified by 2-step PCR, first by using generic primers (step 1) and then by using one generic primer and one index primer to label each sample (step 2). The PCR2 products are then pooled together (library) and analysed by next-generation sequencing (NGS).
Background
Malaria is caused by Plasmodium parasites, which are injected into vertebrate hosts with the bite of a female Anopheles mosquito harbouring the parasite. It is estimated that 282 million cases were recorded in 2024, resulting in the death of 610 thousand people [1]. Reverse genetics has been critical to enable important studies of the parasite’s biology, to ultimately help inform future drug and vaccine intervention and treatment strategies [2]. Plasmodium gene editing, particularly in the human species Plasmodium falciparum, is challenging since the parasites lack the canonical non-homologous end-joining (NHEJ) pathway, have a very AT-rich genome (>80%), and have low transfection efficiency [3]. The rodent malaria species Plasmodium berghei is more genetically tractable, provides a model system to study gene essentiality in vivo, and provides efficient access to mosquito blood and liver stage infection, offering a more complete picture of those parasite genes important throughout the life cycle [4].
Several reverse genetic screens have been performed where a specific group of genes was targeted. This includes the knockout (KO) of all phosphatases and kinases in P. berghei [5,6], the systematic KO of the predicted exportome of P. falciparum [7], and the conditional mis-localisation of all non-secreted proteins on chromosome 3 in P. falciparum [8]. These types of gene-by-gene screens are immensely valuable but extremely time-consuming. More high-throughput KO screens have been completed at different stages of P. berghei (asexual blood stage, sexual commitment, fertilisation, and liver stage), with long-homology arm barcoded vectors from the Plasmodium Genetic Modification (PlasmoGEM) project covering more than half of the protein-coding genome. These barcoded vectors enabled the simultaneous transfection of 100× vectors into mice, where the relative growth of mutant parasites was estimated by tracking mutant barcodes over time using next-generation sequencing (NGS) [9–13]. However, the PlasmoGEM KO vectors could not be generated for the entire protein-coding genome due to technical limitations, and the scaling up of gene tagging vectors had limited success.
The adaptation of clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 gene editing in Plasmodium was done in 2014 [14–17]. In this system, the Cas9 endonuclease introduces a double-strand break at the desired target site, which is determined by the guide RNA (gRNA) sequence. Since Plasmodium does not have the classical NHEJ pathway, the repair is mediated by homologous recombination. Therefore, in addition to the presence of Cas9 and the gRNA, gene edits require a homology-directed repair (HDR) template [18,19]. The parasite also has a microhomology-mediated end-joining pathway, which has been previously used for CRISPR-Cas9 editing; however, this can only be used on genes that harbour repetitive sequences and therefore is not widely used [18,19]. CRISPR has been used successfully for large-scale KO screens in Toxoplasma gondii, another more genetically tractable apicomplexan parasite [20–24]. However, T. gondii has retained the NHEJ pathway, and therefore gene disruption vectors for large KO screens are much easier to generate and do not require the design or supply of an HDR template. Recently, a high-throughput tagging (HiT) system was designed for T. gondii, which relies on homologous recombination to introduce functionalised tags, where the vector contained a gRNA physically linked to the HDR template [25]. This opened the door to scalable CRISPR approaches in parasites that lack the NHEJ pathway.
Here, we present a detailed protocol for the P. berghei high-throughput (PbHiT) CRISPR-Cas9 system to target genes at a scale in P. berghei, inspired by the HiT approach for T. gondii. In our system, short homology arms (100 bp) are physically linked to the gRNA and integrated into the PbHiT vector using a single scalable cloning step. This system can be used for both gene tagging and gene KO, and the vector design can be done manually (as detailed here) or by using our online tool (https://pbhit-crispr-design.serve.scilifelab.se/search). Furthermore, the PbHiT vectors can be pooled together for transfection, where the gRNA serves as an NGS-readable barcode. PbHiT thereby offers the first high-throughput CRISPR system in Plasmodium parasites. Finally, the sequencing data can be analysed and visualised using our scripts freely available at GitHub.
Materials and reagents
Biological materials
1. XL10-Gold ultra-competent cells (Agilent Technologies, catalog number: 200315)
2. pPbU6-hdhfr/yfcu-HiT (Addgene, catalog number: 216421)
3. Other plasmid DNA (Genewiz-Azenta)
4. Plasmodium berghei ANKA Cas9 expressing parasite line (Bushell laboratory, Umeå University) [26]
5. Female BALB/c mice, at least 6 weeks of age (Charles River, Europe)
6. Female Wistar rats >150 g (Charles River, Europe)
Reagents
1. Oligonucleotides (Integrated DNA technologies) (see Table 1)
Table 1. Oligonucleotides used in this protocol
| Name | Sequence (5′ → 3′) |
|---|---|
| PbU6prom_F | ACATATGCGCATACTTCGAGTTATAC |
| hsp70UTR_R | TATGTTCGTGGCATTCCACA |
| BC_pHiT_illumina_F | ACACTCTTTCCCTACACGACGCTCTTCCGATCTCGAATGCACTATTCATTTTATGGGG |
| BC_pHiT_illumina_R | TCGGCATTCCTGCTGAACCGCTCTTCCGATCTGACTCGGTGCCACTTTTTCA |
| PE 1.0 primer | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T |
| Index primers | Sequences can be found in Bushell et al. [9] |
Note: All oligonucleotides except the first two were PAGE-purified.
2. GeneJET Plasmid Miniprep kit (Thermo Fisher Scientific, catalog number: K0502)
3. BsmBI restriction enzyme (New England Biolabs, catalog number: R0580)
4. PstI restriction enzyme (New England Biolabs, catalog number: R0140L)
5. BbsI restriction enzyme (New England Biolabs, catalog number: R0539L)
6. AvrII restriction enzyme (New England Biolabs, catalog number: R0174L)
7. TAE buffer, 50× (VWR, catalog number: K915-4L; 1× TAE made up in distilled water and stored at room temperature)
8. Agarose (VWR, catalog number: 443666A; product format: made up to 0.7% or 1% in 1× TAE buffer)
9. 6× DNA loading dye (New England Biolabs, catalog number: B7024S)
10. T4 DNA ligase (New England Biolabs, catalog number: M0202L)
11. T4 DNA ligase reaction buffer (New England Biolabs, catalog number: B0202S)
12. Macherey-NagelTM NucleoSpinTM Gel and PCR Clean-up (Macherey-Nagel, catalog number: 740609.250)
13. GoTaq G2 Green Master Mix (2×) (Promega, catalog number: M7822)
14. DreamTaq PCR Master Mix (2×) (Thermo Fisher Scientific, catalog number: K1081)
15. SYBR Safe DNA gel stain (Thermo Fisher Scientific, catalog number: S33102)
16. LB broth (Miller) (Sigma-Aldrich, catalog number: L3522; product format: made up in distilled water and autoclaved before use)
17. LB broth with agar (Miller) (Sigma-Aldrich, catalog number: L3147; product format: made up in water and autoclaved before use)
18. Ampicillin (Duchefa, catalog number: A0104, CAS: 69-52-3; product format: 100 mg/mL in water and stored at -20 °C)
19. Kanamycin (Duchefa, catalog number: K016, CAS: 25389-94-0; product format: 50 mg/mL in water and stored at -20 °C)
20. Ethanol (VWR, catalog number: 20821.310)
21. Sodium acetate (Sigma-Aldrich, catalog number: 567422)
22. Methanol (Sigma-Aldrich, catalog number: 34860)
23. Giemsa stain (Sigma-Aldrich, CAS: 51811-82-6; product format: made up to 10% in tap water and stored at room temperature)
24. Phosphate-buffered saline (PBS) (Sigma-Aldrich, Dulbecco, catalog number: D8537-500ML)
25. RPMI-1640 medium, HEPES (Fisher Scientific, Gibco, catalog number: 11594506)
26. Fetal bovine serum (FBS) (Gibco, catalog number: 10500-064)
27. Penicillin-streptomycin (Gibco, catalog number: 15140-122)
28. Sodium bicarbonate (Sigma-Aldrich, catalog number: S5761; product format: 1.2 M made up in distilled water)
29. Histodenz (Sigma-Aldrich, catalog number: D2158-100gr)
30. Heparin (Sigma-Aldrich, catalog number: H4784; product format: 50 mg/mL in Milli-Q water)
31. Pyrimethamine (MP Biomedicals, catalog number: 58-14-0; product format: 0.07 mg/mL in tap water, protect from light)
32. 5-fluorocytosine (Sigma-Aldrich, catalog number: F7129-5G; product format: 1 mg/mL in tap water, protect from light)
33. P3 Primary Cell 4D-Nucleofector X kit S (Lonza, catalog number: V4XP-3032)
34. DNeasy Blood and Tissue kit (Qiagen, catalog number: 69506)
35. Ketaminol (MSD Animal Health, catalog number: 51 15 19, 100 mg/mL)
36. Nerfasin/Xylazine (Dechra Veterinary Products, DIN 02444089, 20 mg/mL)
37. MinElute 96 UF PCR Purification kit (Qiagen, catalog number: 28051)
38. QIAGEN Plasmid Midi kit (Qiagen, catalog number: 12145)
39. Advantage 2 Polymerase mix (TaKaRa, catalog number: 639202)
40. Advantage UltraPure PCR deoxynucleotide mix (TaKaRa, catalog number: 639125)
41. Qubit 1× dsDNA HS Assay kit (Thermo Fisher Scientific, catalog number: Q33231)
42. UltraPureTM DNase/RNase-free distilled water (Fisher Scientific, Invitrogen, catalog number: 11538646)
43. Parasite gas mixture: 1% oxygen, 3% carbon dioxide/nitrogen cylinder (Air Liquide Gas, catalog number: 228447-L)
44. Glycerol (Sigma-Aldrich, catalog number: G5516-100mL)
45. Alsever’s solution (MP Biomedicals, catalog number: 2801154)
46. Hydrochloric acid (Sigma-Aldrich, catalog number: 258148)
Solutions
1. Complete culture medium (see Recipes)
2. Histodenz solution (see Recipes)
3. Terminal anesthetic solution for mice (see Recipes)
4. Terminal anesthetic solution for rats (see Recipes)
5. LB broth with antibiotics (see Recipes)
6. LB agar plates with antibiotics (see Recipes)
7. Glycerol stock (see Recipes)
8. Parasite freezing solution (see Recipes)
Recipes
1. Complete culture medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| RPMI-1640 medium | 72% | 36 mL |
| FBS | 25% | 12.5 mL |
| Sodium bicarbonate | 24 mM | 1 mL of 1.2 M stock |
| Penicillin-streptomycin | 1% | 500 μL |
| Total | 50 mL |
Make fresh before each use.
2. Histodenz solution
To make a Histodenz stock solution (27.6%), first prepare 100 mL of buffered solution and adjust the pH to 7.5, then add the Histodenz (see below). Autoclave the solution (20 min at 120 °C) and store at 4 °C for up to 4 months. Wrap the bottle in aluminium foil to protect it from light.
Stock solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Buffered solution | ||
| Tris-HCl | 5 mM | 60.6 mg |
| KCl | 3 mM | 22.4 mg |
| CaNa2 EDTA | 0.3 mM | 12.3 mg |
| MilliQ water | - | Up to 100 mL total volume |
| Histodenz stock solution | ||
| Buffered solution | - | 100 mL |
| Histodenz | 27.6% | 27.6 g |
Note: Adjust pH using hydrochloric acid (HCl).
Caution: Be careful when handling HCl and follow relevant safety guidelines.
Then, prepare in a 15 mL tube a fresh working solution of Histodenz (15.2%) in PBS (see below), just before using it.
Working solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Histodenz stock solution | 15.2% | 2.75 mL |
| 1× PBS | 45% | 2.25 mL |
| Total | 5 mL |
3. Terminal anesthetic solution for mice
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Ketamine | 33% (33.3 mg/mL) | 2 mL |
| Xylazine | 16.7% (3.33 mg/mL) | 1 mL |
| PBS | 50% | 3 mL |
Make the solution in a 15 mL tube and store at room temperature. The recipe can easily be scaled up if needed. Use a 25 G needle (or bigger) and a 1 mL Luer syringe to draw the solutions through the rubber lid of the glass containers. Do not use past the expiration date of the stock solutions, and always consult with your laboratory animal veterinarian. Weigh mice and use a dose of 225–270 mg/kg of ketamine and 47–60 mg/kg of xylazine.
4. Terminal anesthetic solution for rats
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Ketamine | 66.7% (66.7 mg/mL) | 2 mL |
| Xylazine | 33.3% (6.67 mg/mL) | 1 mL |
Make up the solution in a 15 mL tube and store at room temperature. Use a 25 G needle (or bigger) and a 1 mL Luer syringe to draw the solutions through the rubber lid of the glass containers. Do not use past the expiration date of the stock solutions, and always consult with your laboratory animal veterinarian. Weigh rats and use a dose of 225–270 mg/kg of ketamine and 15–30 mg/kg of xylazine.
5. LB broth with antibiotics
Dilute ampicillin or kanamycin stock 1:1,000 in LB broth before use.
6. LB agar plates with antibiotics
Melt the LB agar completely by placing the bottle in a water bath (42 °C) or in a microwave. Once melted, wait for the agar to cool down to ~50 °C before adding ampicillin or kanamycin (1:1,000 dilution), mix gently, and pour onto plates (enough to cover the surface area). Let agar settle at room temperature; once solid, stack plates together and store in the fridge (up to approximately 1 month).
7. Glycerol stock
Make up a 50% glycerol stock solution in Milli-Q water and autoclave. Store at room temperature. Mix 1:1 bacteria culture and glycerol stock solution to make up a 25% glycerol stock. Store bacteria glycerol stock at -80 °C.
8. Parasite freezing solution
Mix in a 50 mL tube 5 mL of 100% glycerol and 45 mL of Alsever’s solution. Mix well and filter-sterilise using a 0.2 μm filter. Store 10 mL aliquots at -20 °C for long-term storage or at 4 °C for short-term storage.
Laboratory supplies
1. 1.5 mL safe-lock tubes (Fishes Scientific, Eppendorf, catalog number: 15625367)
2. Screw cap tube, 15 mL (Sarstedt, catalog number: 62.554.502)
3. Screw cap tube, 50 mL (Sarstedt, catalog number: 62.547.254)
4. Falcon polypropylene round-bottom test tubes (loose cap) (Merck, Corning, catalog number: CLS352006)
5. No. 23 scalpel blade (Scientific Laboratory Supplies, Swann-Morton, catalog number: INS4688)
6. Microscope slides (Thermo Scientific, Epredia, catalog number: 16247920)
7. Narrow mouth Erlenmeyer flask, reusable (Merck, Corning, catalog number: CLS4985P500)
8. BD PlastiPakTM Luer slip syringe without needle (Thermo Fisher, catalog number: 15489199)
9. 25 G needles (Fisher Scientific, BD Microlance, catalog number: 10442204)
10. Omnican 20 U insulin injection syringe (Fisher Scientific, B Braun, catalog number: 15188928)
11. Sterile blood lancets (Vitrex Medical, catalog number: 357213)
12. Benchtop cooler (Thermo Scientific, catalog number: 10777232)
13. Polypropylene micro tube rack, pack of 5 (Thermo Fisher, catalog number: 11728084)
14. Delrin full-size test tube racks, 24 mm × 30 mm (Thermo Fisher, catalog number: 10257963)
15. 1.8 mL cryotubes (Thermo Scientific, catalog number: 368632)
16. 5 mL serological pipettes (VWR, catalog number: 612-3702)
17. 10 mL serological pipettes (VWR, catalog number: 612-3700)
18. 25 mL serological pipettes (VWR, catalog number: 612-3698)
19. PCR tubes (VWR, catalog number: 732-0545)
20. 10 μL tips (VWR, catalog number: 613-6463)
21. Tips 200 μL sterile filtered tips (VWR, catalog number: 732-3397)
22. 1,000 μL tips (VWR, catalog number: 613-6471)
23. 90 mm plates (Thermo Scientific, catalog number: 101VR20)
24. Spreader, T-shaped (VWR, catalog number: 612-2651)
25. Reagent reservoirs (VWR, catalog number: 613-1181)
26. 96-well deep-well plate (VWR, catalog number: 732-2893)
27. Pasteur pipette (VWR, catalog number: 612-1681)
28. PCR microplate (Axygen, catalog number: PCR-96-LP-AB-C)
29. Microplate sealing film, rayon (Fisher Scientific, Axygen, catalog number: 11326254)
30. Microplate sealing film, aluminium (Fisher Scientific, Axygen, catalog number: 12577947)
31. MillexTM-GS filter unit (sterile) (Merck, Millipore, catalog number: SLGS033)
Equipment
1. Eppendorf Research plus, 4-pack (Eppendorf, catalog number: 3123000950)
2. Pipetboy acu 2 (VWR, catalog number: 612-2964)
3. Beckman Coulter centrifuge (Beckman Coulter, model: Allegra V-15R, catalog number: C63126)
4. PicoTM 21 microcentrifuge (Thermo Fisher Scientific, catalog number: 75002553)
5. C1000 TouchTM thermal cycler with 96-well fast reaction module (Bio-Rad, catalog number: 1851196)
6. Eppendorf ThermoMixer C (Merck, Eppendorf, catalog number: EP5382000015)
7. Eppendorf ThermoTop (Merck, Eppendorf, catalog number: EP5308000003)
8. IKA KS 4000 i incubator shaker (Fisher Scientific, IKA, catalog number: 12944035)
9. Benchtop orbital shaker (Eppendorf, New Brunswick, model: Innova® 40, catalog number: M1299-0092)
10. DUALED blue/white transilluminator (Bioneer, catalog number: A-6020)
11. ChemiDoc imaging system (Bio-Rad, catalog number: 12003153)
12. Amaxa 4D-NucleofectorTM core unit (Lonza, catalog number: AAF-1002B)
13. D-Nucleofector® X unit (Lonza, catalog number: AAF-1003X)
14. Counting chamber with double net ruling and V-slash (Fisher Scientific, Marienfeld Superior, catalog number: 15110535)
15. UVP PCR3 HEPA workstation (analytik jena, catalog number: 849-95-0600-02)
16. QIAvac connecting system (Qiagen, catalog number: 19419)
17. QIAvac 96 (Qiagen, catalog number: 19504)
18. Mice incubator (Vet-Tech, VTS Serial No. 1458, Code No. HEO11)
19. NGS sequencing platform (e.g., Illumina MiSeq, NextSeq, NovaSeq)
20. Nanodrop (IMPLEN, catalog number: T81946)
21. Lab bench (DanLaf VFRS 1206, Scanbur, catalog number: 5522190A) and suction system N840 Laboport (KNF, catalog number: 5220 19 0612, Hose connector, catalog number: 317278, key for hose connector, catalog number: 316279, spare parts kit, catalog number: N 840 G 317436)
22. PCR roller (Merck, catalog number: AXYPCRSPROLLER)
23. 8-channel multichannel pipette (Eppendorf, catalog number: 3125000036)
Software and datasets
1. EuPaGDT tool: http://grna.ctegd.uga.edu/ [27] EuPaGDT: a web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens, free
2. PlasmoDB: https://plasmodb.org/plasmo/app [28] VEuPathDB: the eukaryotic pathogen, vector, and host Bioinformatics Resource Center, free
3. Benchling (Biology software): 2024, https://www.benchling.com/, free
4. NEBio Calculator (New England Biolabs, v1.15 and v1.16): https://nebiocalculator.neb.com/, free
5. Image Lab Software, Bio-Rad (Version 6.1) (SOFT-LIT-170-9690-ILSPC-V-6-1), free
6. R: https://www.r-project.org/ [29], free
7. RStudio: http://www.posit.co/ [30], free
8. PbHiT data analysis and visualisation pipeline: https://github.com/henriksson-lab/pbhit_analyzer, free
9. R package minpack.lm v.1.2–4: https://CRAN.R-project.org/package=minpack.lm [31] minpack.lm: R Interface to the Levenberg-Marquardt Nonlinear Least-Squares Algorithm Found in MINPACK, Plus Support for Bounds. R package
10. RShiny: https://cran.r-project.org/web/packages/shiny/index.html [32] shiny: Web Application Framework for R. R package version 1.9.0.
11. ggplot2: https://ggplot2.tidyverse.org [33] ggplot2: Elegant Graphics for Data Analysis.
12. plotly: https://plotly-r.com [34] Interactive Web-Based Data Visualisation with R, plotly, and shiny.
13. PbHiT online design tool: https://pbhit-crispr-design.serve.scilifelab.se/search [26].
Procedure
文章信息
稿件历史记录
提交日期: Sep 16, 2025
接收日期: Dec 9, 2025
在线发布日期: Dec 29, 2025
出版日期: Jan 20, 2026
版权信息
© 2026 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Jonsdottir, T. K., Paoletta, M. S., Henriksson, J. and Bushell, E. S. (2026). Plasmodium berghei High-Throughput (PbHiT): a CRISPR-Cas9 System to Study Genes at Scale. Bio-protocol 16(2): e5572. DOI: 10.21769/BioProtoc.5572.
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link









