- EN - English
- CN - 中文
Correcting Image Distortion in Expansion Microscopy Using 3D-Aligner
发布: 2026年01月20日第16卷第2期 DOI: 10.21769/BioProtoc.5568 浏览次数: 61
评审: Olga KopachAnonymous reviewer(s)
Abstract
Expansion microscopy (ExM) is an innovative and cost-effective super-resolution imaging technique that enables nanoscale visualization of biological structures using conventional fluorescence microscopes. By physically enlarging biological specimens, ExM circumvents the diffraction limit and has become an indispensable tool in cell biology. Ongoing methodological advances have further enhanced its spatial resolution, labeling versatility, and compatibility with diverse sample types. However, ExM imaging is often hindered by sample drift during image acquisition, caused by subtle movements of the expanded hydrogel. This drift can distort three-dimensional reconstruction, compromising both visualization accuracy and quantitative analysis. To overcome this limitation, we developed 3D-Aligner, an advanced and user-friendly image analysis software that computationally corrects sample drift in fluorescence microscopy datasets, including but not limited to those acquired using ExM. The algorithm accurately determines drift trajectories across image stacks by detecting and matching stable background features, enabling nanometer-scale alignment to restore structural fidelity. We demonstrate that 3D-Aligner robustly corrects drift across ExM datasets with varying expansion factors and fluorescent labels. This protocol provides a comprehensive, step-by-step workflow for implementing drift correction in ExM datasets, ensuring reliable three-dimensional imaging and quantitative assessment.
Key features
• 3D-Aligner precisely corrects sample drift in expansion microscopy (ExM) datasets, enabling reliable 3D reconstruction and robust quantitative analysis.
• Utilizes background feature detection and feature matching across z-planes to achieve nanoscale-precision drift correction.
• 3D-Speckler, which is a MATLAB-based software platform, offers a customizable and user-friendly interface.
• Outperforms conventional registration tools across varying expansion factors and labeling conditions and is equally applicable to non-ExM datasets.
Keywords: Expansion microscopy (ExM)Background
Fluorescence microscopy has become a cornerstone technique in biological research, enabling visualization of molecular structures and dynamic processes within cells and tissues. However, its optical resolution is fundamentally limited to approximately 250 nm laterally and 500 axially [1,2], leaving many subcellular structures unsolved. Although advanced super-resolution methods such as STED, SIM, STORM, and PALM have been developed to overcome this limitation, they often require specialized instrumentation, complex sample preparation, and intensive computational processing [1,3–5].
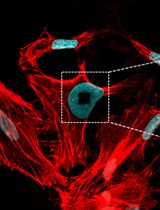
Expansion microscopy (ExM) provides a cost-effective and accessible alternative by physically enlarging biological specimens embedded in a swellable polyacrylamide-based hydrogel [5–7]. ExM improves the effective resolution in proportion to the expansion factor, enabling nanoscale visualization of cellular and tissue architecture using standard fluorescence microscopes [6,8–10]. Despite its simplicity and versatility, ExM imaging remains challenged by sample drift, primarily caused by subtle movements of the soft, water-rich hydrogel during three-dimensional (3D) image acquisition. Such drift introduces spatial distortions that degrade image quality and compromise the accuracy of quantitative analyses, particularly in large or highly expanded samples [7,11–13].
A variety of experimental approaches, such as poly-L-lysine coating, agarose embedding, and mechanical stabilization, have been explored to minimize drift. However, these methods often yield inconsistent results or introduce undesirable artifacts, including sample shrinkage or fluorescence loss [11–14]. Consequently, post-acquisition drift correction has emerged as a more reliable and noninvasive strategy to ensure accurate 3D reconstruction and quantitative measurement.
Existing software tools, including several ImageJ plugins [15,16], provide limited functionality for drift or distortion correction in ExM datasets. Most were originally designed for time-lapse image registration rather than z-stack alignment, and thus often require metadata modification, manual parameter tuning, or correlation-based alignment methods that lack the precision needed for quantitative ExM analysis.
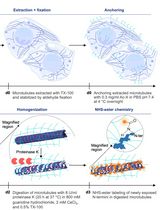
To address this gap, we developed 3D-Aligner, a dedicated computational tool optimized for drift correction in 3D fluorescence microscopy datasets, and validated its performance, particularly using ExM images [14]. 3D-Aligner is fully integrated into our previously developed 3D-Speckler platform, designed for nanometer-scale particle analysis in fluorescence microscopy [2,17]. The 3D-Aligner software detects faint, nonspecific background fluorescence signals and applies a nearest-neighbor matching algorithm between adjacent z-planes to determine drift trajectories with nanometer precision. By relying on background features rather than biological structures, 3D-Aligner minimizes correction-induced artifacts and ensures unbiased alignment.
The tool offers a streamlined, user-friendly interface with customizable parameters for feature size, expected drift range, and channel selection. Users can visualize drift trajectories and corrected image stacks to facilitate downstream quantitative analysis. Benchmarking results demonstrate that the 3D-Aligner outperforms existing registration programs in both correction accuracy and reconstruction fidelity across diverse expansion factors and labeling conditions [14].
Beyond ExM, the 3D-Aligner can be applied to particle tracking, motion analysis, or distortion correction in other fluorescence microscope methods. We also demonstrated its capability to perform nanoscale particle tracking in live-cell imaging datasets. Overall, the 3D-Aligner provides a robust, accessible, and precise solution for drift correction, significantly enhancing the reliability of quantitative 3D imaging in expansion microscopy and related super-resolution methods. In this protocol, we present a detailed, step-by-step guide for configuring and using 3D-Aligner, illustrated with example drifted datasets from 4-fold expanded samples using the 4×3D-ExM method, which represents one of the most commonly used ExM approaches in a wide range of research fields [6].
Software and datasets
1. MATLAB (MathWorks, R2019-b and above, 10/1/2018, license required)
Note: For users who do not have access to a MATLAB license, we provide a standalone, executable application made through MATLAB compiler. The standalone 3D-Speckler (including the 3D-Aligner module) and user manual are available at the following website (Windows: https://drive.google.com/drive/folders/1om5kwV7qFZvelzwLzCHVQVjJ2BhD6Gts?usp=sharing and Mac: https://drive.google.com/drive/folders/1gaFIiaPas6SSCpkxSLYmIKGqqWjl1fpI?usp=sharing).
2. 3D-Speckler (including the 3D-Aligner module) (https://github.com/suzukilabmcardle/3D-Speckler or https://drive.google.com/drive/folders/15_nXhPmW60pvuuSI_lxOB-VsXEWtwGwD) (07/20/2024, publicly available) (see General note 1)
Procedure
文章信息
稿件历史记录
提交日期: Nov 6, 2025
接收日期: Dec 11, 2025
在线发布日期: Dec 25, 2025
出版日期: Jan 20, 2026
版权信息
© 2026 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Hsiao, W. Y., Ghone, D. and Suzuki, A. (2026). Correcting Image Distortion in Expansion Microscopy Using 3D-Aligner. Bio-protocol 16(2): e5568. DOI: 10.21769/BioProtoc.5568.
分类
生物工程
细胞生物学 > 细胞成像 > 超分辨率成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link