- EN - English
- CN - 中文
Efficient Fluorescent Labeling of Human Trophoblast Stem Cells via a CRISPR/Cas9-Mediated Knock-In Approach in a Safe Harbor Locus
发布: 2026年01月05日第16卷第1期 DOI: 10.21769/BioProtoc.5561 浏览次数: 51
评审: Rajesh RanjanAnonymous reviewer(s)
Abstract
Labeling cells with reporter genes allows researchers to visually identify specific cells and observe how they interact with each other in dynamic biological systems. Even though various labeling methods are now available, a specific description of gene knock-in labeling methods for human trophoblast stem cells (hTSCs) has not been reported. Here, we present a streamlined protocol for labeling hTSCs with the green fluorescent protein (GFP) reporter gene via CRISPR/Cas9-mediated knock-in of the gene into the adeno-associated virus site 1 (AAVS1) safe harbor locus. A commonly used hTSC cell line, CT29, was transfected with a dual plasmid system encoding the Cas9 endonuclease and an AAVS1-targeted guide RNA in one plasmid and a donor plasmid encoding a puromycin resistance gene and GFP reporter gene flanked by AAVS1 homology arms. Puromycin-resistant clonal cells were isolated, and AAVS1 integration was confirmed via PCR and sequencing of the PCR products. The labeled cells are proliferative and can give rise to extravillous cytotrophoblast cells (EVT) and the syncytiotrophoblast (ST). To our knowledge, this is the first report using the CRISPR/Cas9 system for AAVS1 integration of a reporter gene in human trophoblast stem cells. It provides an efficient tool to facilitate the study of human trophoblast development and function in co-culture systems and will be highly useful in developing clinical gene therapy-related plasmid constructs.
Key features
• First report to constitutively express a fluorescent label in hTSCs by applying a CRISPR/Cas9 knock-in approach and an AAVS1 safe harbor locus.
• Provides an efficient tool to facilitate the study of human trophoblast development and function, particularly in heterologous co-culture systems.
• Offers an approach for developing clinical gene therapy–related plasmid constructs that allow insertion of therapeutic genes without associated disruption of essential genes.
• Widely applicable approach to label other human cell lines.
Keywords: hTSCsGraphical overview
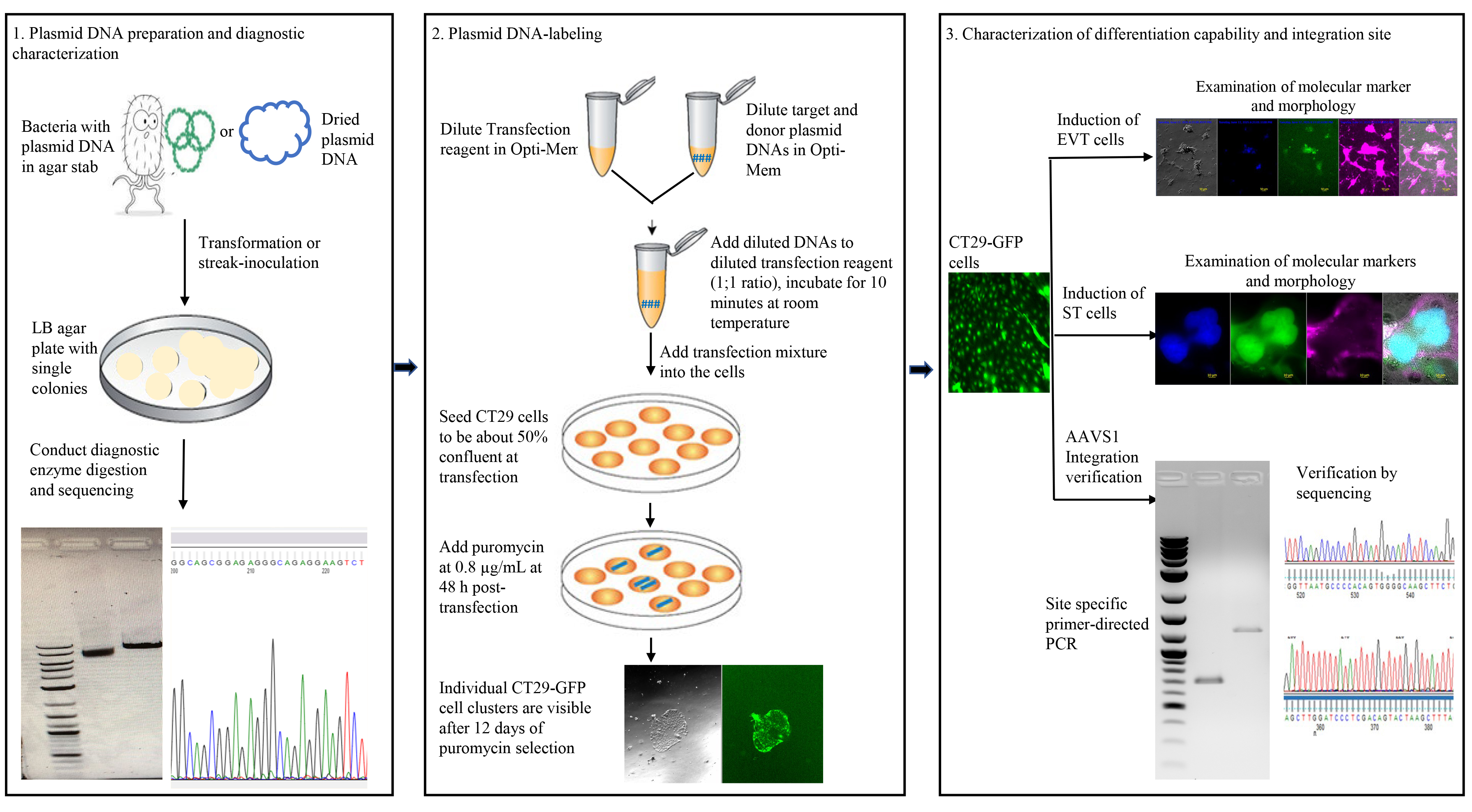
Background
Placental trophoblast cells are formed during the earliest stages of pregnancy and play a pivotal role in the complex interactions between the fetus and the mother. These early stages of human pregnancy are difficult to study due to inherent biological as well as legal and ethical factors; as such, in vitro modeling of human embryo implantation and placentation is necessary. In the human placenta, three major trophoblast subpopulations exist, which include cytotrophoblast cells (CT), extravillous cytotrophoblast cells (EVT), and the syncytiotrophoblast (ST) [1–3]. CT cells are capable of self-renewal and represent the undifferentiated progenitors of EVT and ST. Differentiated trophoblast types interact with many maternal and fetal cells types, including endometrial epithelial and stromal cells, maternal decidual macrophages, uterine natural killer cells, regulatory T cells, and fetal Hofbauer cells [4–6]. Coordinated proliferation and differentiation of trophoblast cells and appropriate interactions with these other cells and tissues are vital for a successful pregnancy. Impaired trophoblast development and function can lead to various pregnancy complications, including miscarriage, pre-eclampsia, and intrauterine growth restriction [7].
Labeling cells with reporter genes like the green fluorescent protein (GFP) gene allows researchers to easily identify cells under a microscope, making such labeling a crucial tool to study multicell interactions and to track the fate of specific cell populations during development or in response to stimuli. This is particularly useful when studying trophoblast development and placentation in vitro, and more specifically in heterologous cell co-culture models representing embryo implantation. Reporter gene cell labeling commonly involves integrating the reporter genes into the genomes of the cells to be studied in order to stably produce the reporter proteins throughout the lifetime of the cell and in any subsequent daughter cells. Over the years, many labeling methods have been developed, each with its own strengths, weaknesses, and specific applications. For instance, lentiviral vectors have often been used for transgene integration because they can carry large transgenes and integrate into both dividing and non-dividing cells, leading to stable and long-term expression. However, lentiviral vectors insert their genetic material at multiple random locations within the host genome, which has the potential to cause unwanted off-target effects such as insertional mutagenesis or oncogene activation [8]. Zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) are both well-established protein-based gene editing tools, but their design and construction are time-consuming, expensive, and challenging, as unique nuclease sequences must be generated for every genomic target. Alternatively, use of clustered regularly interspaced short palindromic repeats/Cas9 (CRISPR/Cas9), which was developed by several groups in 2013, has revolutionized gene editing by offering a method that is quicker, cheaper, and easier to design than previous approaches [9–11]. Importantly, CRISPR/Cas9 has also been shown to have higher efficiency and high specificity when compared to previously used gene editing tools, making it a promising approach for targeted integration of reporter genes [9–11].
Human trophoblast stem cells (hTSCs) are a specialized stem cell type that can be challenging to label using genetic tools because they are not robustly proliferative in culture and require a highly controlled microenvironment, including specific combinations of growth factors and signaling pathway modulators, to prevent differentiation in vitro. Further, their reluctance to grow under sparse culture densities makes certain genetic engineering methods that require cloning particularly difficult. While various cell-labeling techniques are available, and the CRISPR/Cas9-based approach has been used to perform gene knockout and transcriptional activation in hTSCs [12], there are few published studies specifically detailing the application of gene knock-in labeling methods in hTSCs. As a part of our broader and in-depth studies, we have developed a protocol to label hTSCs with the GFP reporter gene via CRISPR/Cas9-mediated knock-in into the adeno-associated virus site 1 (AAVS1). Here, AAVS1 lies within the first intron of the constitutively expressed PPP1R12C gene and acts as an important safe harbor where exogenous pieces of DNA can integrate and function in a predictable manner without causing adverse and unpredictable changes to the host genome or substantial risk to the host cell [13–15]. The resultant labeled hTSC line is an efficient tool to facilitate the study of human trophoblast development and function. Furthermore, this knock-in genome editing system should prove highly useful in a wide range of preclinical and potentially clinical gene therapy studies by offering a reliable method to develop genetically engineered plasmid constructs that can be introduced into human trophoblast progenitors.
Materials and reagents
Biological materials
1. CT29, a human trophoblast stem cell line derived from first-trimester placental cytotrophoblast cells (RIKEN BRC cell bank, catalog number: RCB4937), kindly provided by Dr. Okae [1] and Dr. Liping Feng at Duke University
Note: It is often recommended to use low-passage cells. The passage numbers we used for the CT29 cell line were between 19 and 25, and mycoplasma testing was performed periodically.
2. pCas-Guide-AAVS1, an all-in-one AAVS1 gRNA plasmid, delivered as a lyophilized 10 μg product (Origene, catalog number: GE100023)
3. pAAVS1-P-CAG-GFP, a donor plasmid for AAVS1 targeting and constitutive GFP expression, delivered as a bacterial agar slab (Addgene, catalog number: 80491)
Note: These are both high-copy plasmids with typical yields being 10–30 μg from a 1–5 mL bacterial culture volume when purified with a plasmid miniprep kit. We used DH5α competent cells to do the routine transformation.
4. 3′-junction directed primers: 5′-TCATTGCAATAGTGTGTTGG-3′ and 5′-AGCGGCCGCGAATTCGCCCTTAG-3′
5. 5′-junction directed primers:5′-CTGCACCACGTGATGTCCTCTG-3′ and 5′-GTGGGCTTGTACTCGGTCATC-3′
Note: These primers were designed by using the Oligonucleotide Properties Calculator (see Software) and were ordered from Integrated DNA Technologies. Standard desalting was performed by the vendor to purify them.
Reagents
1. DMEM/F12 (Thermo Fisher Scientific, catalog number: 11320033)
2. ITS-X (Thermo Fisher Scientific, catalog number: 51500056)
3. CHIR-99021 (Reprocell, catalog number: 04-0004); prepare at 3 mM in DMSO and store at -20 °C as 533 μL aliquots
4. Y-27632 (Reprocell, catalog number: 04-0012-02); prepare at 10 mM in MBG water and store at -20 °C as 50 μL aliquots
5. SB431542 (Reprocell, catalog number: 04-0010); make 10 mM SB431542 in DMSO and store as 80 μL aliquots at -20 °C
6. A83-01 (Reprocell, catalog number: 04-0014); make 5 mM A83-01 in DMSO and store as 80 μL aliquots at -20 °C
7. Valproic acid (VPA) (Sigma-Aldrich, catalog number: P4543); prepare 300 mM in sterile MBG water and store at 4 °C after aliquoted
8. KnockOut serum replacement (KOSR) (Thermo Fisher Scientific, catalog number: 10828010)
9. NRG1-beta 1, human (MCE, catalog number: HY-P7365); prepare at 200 μg/mL in 0.2% FBS in PBS and store in 25 μL aliquots at -20 °C
10. Collagen IV mouse (Fisher Scientific, catalog number: CB-40233)
Note: Collagen IV should be stored at -20 °C, and handling should always occur on ice. Collagen IV can be thawed by placing it on ice and slowly mixing it with an ice-cold buffer under sterile conditions.
11. BSA (Sigma-Aldrich, catalog number: A7906)
12. FBS (Sigma-Aldrich, catalog number: F2442)
13. EGF, human (hEGF) (Sigma-Aldrich, catalog number: E9644); dissolve in 0.2% FBS in PBS at 100 μg/mL and then aliquot at 250 μL and store at -20 °C
14. Foskolin (Sigma-Aldrich, catalog number: F6886); store powder at room temperature; store 5 mM DMSO solution at -20 °C as 500 μL aliquots
15. 2-Mercaptoethanol (2-ME) (Thermo Fisher Scientific, catalog number: 21985-023)
16. L-Ascorbic acid 2-phosphate sesquimagnesium salt hydrate (Sigma-Aldrich, catalog number: A8960); prepare 30 mg/mL in MBG water and store at -20 °C after aliquoted
17. Penicillin-streptomycin (Pen/Strep, 10,000 U/mL) (Thermo Fisher Scientific, catalog number: 15140122)
18. Matrigel matrix (Fisher Scientific, catalog number: CB-40234C)
Note: Matrigel should be stored at -20 °C, and handling should always occur on ice. To thaw, place Matrigel overnight in a 2–8 °C ice bath or 4 °C refrigerator. Always use chilled labware and tips, keep Matrigel on ice, and aliquot after the first thaw to minimize freeze-thaw cycles. Matrigel is not growth factor-reduced (GFR) and contains several growth factors.
19. Certified molecular biology agarose (Bio-Rad, catalog number: 1613101)
20. Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418)
21. 10× phosphate-buffered saline (PBS), without calcium and magnesium (Corning, catalog number: 46-013-CM)
22. 10× TAE buffer (Corning, catalog number: 46-010-CM)
23. Molecular biology grade (MBG) water (Corning, catalog number: 46-000-CM)
24. HyClone cell culture grade water (Cytiva, catalog number: SH30529.02)
25. HyClone Dulbecco's phosphate-buffered saline (DPBS) without calcium, magnesium (Cytiva, catalog number: SH30028-03)
26. Miller’s LB broth (Corning, catalog number: 46-050-CM)
27. TrypLE Express enzyme (Thermo Fisher Scientific, catalog number: 12604-021)
28. Lipofectamine Stem transfection reagent (Thermo Fisher Scientific, catalog number: STEM00008)
29. Opti-MEM 1 (Thermo Fisher Scientific, catalog number: 31985062)
30. CELLBANKER 1, ready-to-use serum-containing cell cryopreservation medium (Amsbio, catalog number: 11888)
Note: The volumes of the aliquots made for the reagents above are usually just enough to make the inhibitor cocktail or the complete medium for routine use (see Recipes). Frequent freeze-thaw cycles should be avoided when handling these reagents.
Solutions
1. Complete trophoblast stem cell (TS) growth media (see Recipes)
a. Basal media
b. Inhibitor cocktail
2. EVT differentiation medium (see Recipes)
a. Basal differentiation media
b. Initial differentiation media
c. Day 3 differentiation media
d. Day 6 differentiation media
3. ST (2D) differentiation medium (see Recipes)
a. Basal differentiation media
b. ST 2D complete differentiation media
Recipes
1. Complete trophoblast stem cell (TS) growth medium
Adapted from Okae et al. for routine use [1]. This medium is composed of basal medium, inhibitor cocktail, VPA, and Y-27632 stock solutions, and the recipes are shown below.
a. Basal media (500 mL)
| Reagent | Final concentration | Volume |
|---|---|---|
| DMEM/F12 | n/a | 486 mL |
| BSA, 30% prepared in sterile MBG water | 0.3% | 5 mL |
| ITS-X, 100× | 1% | 5 mL |
| FBS | 0.2% | 1 mL |
| Pen/strep (optional) (10,000 U/mL) | 0.5% | 2.5 mL |
| 2-ME, 55 mM | 0.1 mM | 900 μL |
| hEGF, 100 μg/mL | 50 ng/mL | 250 μL |
| L-ascorbic acid, 30 mg/mL | 1.5 μg/mL | 25 μL |
Store at 4 °C and use within 6 months.
b. Inhibitor cocktail (800 μL)
| Reagent | Final concentration | Volume |
|---|---|---|
| 3 mM CHIR-99021 | 2 mM | 533 μL |
| 5 mM 83-01 | 0.5 mM | 80 μL |
| 10 mM SB431542 | 1 mM | 80 μL |
| DMSO | n/a | 107 μL |
Store at -20 °C and use within 6 months.
c. Complete TS growth media (10 mL)
| Reagent | Final concentration | Volume |
|---|---|---|
| Basal media | n/a | 10 mL |
| Inhibitor cocktail | n/a | 10 μL |
| 300 mM VPA | 0.8 mM | 26.6 μL |
| 10 mM Y-27632 | 5 μM | 5 μL |
Store at 4 °C and use within 2 weeks.
2. EVT differentiation medium
Adapted from Okae et al. [1]. Medium compositions for performing EVT differentiation at different time points are slightly different. The recipes are shown below.
a. Basal differentiation media (50 mL)
| Reagent | Final concentration | Volume |
|---|---|---|
| DMEM/F12 | n/a | 48.2 mL |
| BSA, 30% | 0.3% | 0.5 mL |
| ITS-X, 100× | 1% | 0.5 mL |
| Pen/strep (optional) (10,000 U/mL) | 0.5% | 0.25 mL |
| 2-ME, 55 mM | 0.1 mM | 91 μL |
| A83-01, 5 mM | 7.5 μM | 75 μL |
| Y-27632, 10 mM | 2.5 μM | 12.5 μL |
Store at 4 °C and use within 6 months.
b. Initial differentiation media (12 mL)
| Reagent | Final concentration | Volume |
|---|---|---|
| Basal differentiation media | n/a | 12 mL |
| KnockOut serum replacement | 4% | 480 μL |
| NRG1-beta 1 | 100 ng/mL | 6 μL |
| Matrigel matrix (see the note below) | 2% | 240 μL |
Make fresh and prepare on demand.
c. Day 3 differentiation media
| Reagent | Final concentration | Volume |
|---|---|---|
| Basal differentiation media | n/a | 12 mL |
| KnockOut serum replacement | 4% | 480 μL |
| Matrigel matrix (see the note below) | 0.5% | 60 μL |
Make fresh and prepare on demand.
d. Day 6 differentiation media
| Reagent | Final concentration | Volume |
|---|---|---|
| Basal differentiation media | n/a | 12 mL |
| Matrigel matrix (see the note below) | 0.5% | 60 μL |
Make fresh and prepare on demand.
Note: Other required components should be added first in a tube and keep the tube on ice for a few minutes, add Matrigel last to the mixture, and then mix gently and quickly before plating.
3. ST (2D) differentiation medium
Adapted from Okae et al. [1]. This medium is composed of basal differentiation media, KOSR, Y-27632, and forskolin stock solutions, and the recipes are shown below.
a. Basal differentiation media (500 mL)
| Reagent | Final concentration | Volume |
|---|---|---|
| DMEM/F12 | n/a | 486 mL |
| BSA, 30% | 0.3% | 5 mL |
| ITS-X, 100× | 1% | 5 mL |
| Pen/strep (optional) (10,000 U/mL) | 0.5% | 2.5 mL |
| 2-ME, 55 mM | 0.1 mM | 900 μL |
Store at 4 °C and use within 6 months.
b. ST 2D complete differentiation media (12 mL)
| Reagent | Final concentration | Volume |
|---|---|---|
| Basal differentiation medium | n/a | 12 mL |
| Y-27632, 10 mM | 2.5 μM | 3 μL |
| KnockOut serum replacement | 4% | 480 μL |
| Forskolin, 5 mM | 2 μM | 4.8 μL |
Make fresh and prepare on demand.
Laboratory supplies
1. 8-strip PCR tubes and caps (Genesee Scientific, catalog number: 24-705)
2. 1.5 mL tubes (Sigma-Aldrich, catalog number: HS4323)
3. 15 mL and 50 mL disposable centrifuge tubes (VWR, catalog numbers: 89039-664 and 89039-660)
4. 16 G BD precision Glide needle (BD, catalog number: 305198)
5. 10 mL syringe (BD, catalog number: 309604)
6. 50 mL syringe (BD, catalog number: 309653)
7. 0.22 μm, 30 mm, PVDF syringe filter (Vista Lab Technologies, catalog number: 229743)
8. Corning® cell strainer, pore size 40 μm (Corning, Millipore Sigma, catalog number: CLS431750-50EA)
9. 250 mL and 500 mL filter systems (Corning, catalog numbers: 431096 and 430769)
10. VWR® disposable Petri dishes, 60 × 15 mm, mono Petri dishes (VWR International, catalog number: 25384-168)
11. Snap Cap low retention microcentrifuge tubes (Thermo Fisher Scientific, catalog number: 3448PK)
12. Adjustable volume pipettes: P100–1000 μL, P10–100 μL, P2–20 μL, and P0.5–10 μL
Equipment
1. Nanodrop 2000/2000C spectrophotometer (Thermo Fisher Scientific, catalog number: ND2000CLABTOP)
2. Gel electrophoresis assembly (Bio-Rad, model: 200/2.0)
3. Corning® Axygen® refrigerated microcentrifuge (Corning, Millipore Sigma, catalog number: AXY60105021)
4. Beckman Coulter Microfuge 18 centrifuge (Fisher Scientific, catalog number: NC2380892)
5. CO2 incubator for cell culture (Thermo Fisher Scientific, model: 3310)
6. Eppendorf 5331 MasterCycler gradient thermal cycler
7. Zeiss Axio Observer Z1 inverted fluorescence motorized microscope (Zeiss, catalog number: SM100331-MG1MOTC); objective type: EC Plan-Ncofluar 10×/0.30 Ph 1; filters: 450–490, 500–550
8. Sanger DNA sequencing facility (GENEWIZ, https://www.genewiz.com)
9. Automated cell counter (Fisher Scientific, catalog number: AMQAX2000)
Software and datasets
1. ZEN software for Carl Zeiss fluorescence microscope (Zen 2.3, blue edition, v2.3.69.1000): used to analyze all the fluorescent imaging pictures
2. Oligonucleotide Properties Calculator: used to design the primers (https://www.biosyn.com/gizmo/tools/oligo/oligonucleotide%20properties%20calculator.htm)
3. FinchTV (https://finchtv.software.informer.com): used to view the sequencing data
Procedure
文章信息
稿件历史记录
提交日期: Oct 10, 2025
接收日期: Dec 2, 2025
在线发布日期: Dec 25, 2025
出版日期: Jan 5, 2026
版权信息
© 2026 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Zhang, H., Zhou, J., Orsolini, M., Zhao, A., Takhirov, A. and Schust, D. J. (2026). Efficient Fluorescent Labeling of Human Trophoblast Stem Cells via a CRISPR/Cas9-Mediated Knock-In Approach in a Safe Harbor Locus. Bio-protocol 16(1): e5561. DOI: 10.21769/BioProtoc.5561.
分类
干细胞 > 胚胎干细胞 > 基于细胞的分析方法
分子生物学 > DNA > 染色体工程
生物科学 > 生物技术 > CRISPR/Cas9
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link