- EN - English
- CN - 中文
SiMPull-POP: Quantification of Membrane Protein Assembly via Single Molecule Photobleaching
SiMPull-POP:利用单分子光漂白技术定量分析膜蛋白的组装
发布: 2026年01月05日第16卷第1期 DOI: 10.21769/BioProtoc.5560 浏览次数: 280
评审: Olga KopachAnonymous reviewer(s)

相关实验方案
Cell-Sonar:通过特定蛋白标志物表达变化追踪目标蛋白的简便低成本方法
Sabrina Brockmöller [...] Simone Rothmiller
2025年02月05日 1654 阅读
Abstract
Traditional methods for studying protein–protein interactions often lack the resolution to quantitatively distinguish distinct oligomeric states, particularly for membrane proteins within their native lipid environments. To address this limitation, we developed SiMPull-POP (single-molecule pull-down polymeric nanodisc photobleaching), a single-molecule technique designed to quantify membrane protein oligomerization with high sensitivity and in a near-native context. The goal of SiMPull-POP is to enable precise, quantitative analysis of membrane protein assembly by preserving native lipid interactions using diisobutylene maleic acid (DIBMA) to form nanodiscs. Unlike ensemble methods such as co-immunoprecipitation or FRET, which average out heterogeneous populations, SiMPull-POP uses photobleaching to resolve monomeric, dimeric, and higher-order oligomeric states at the single-molecule level. We validated SiMPull-POP using several model systems. A truncated, single-pass transmembrane protein (Omp25) appeared primarily monomeric, while a membrane-tethered FKBP protein exhibited ligand-dependent dimerization upon addition of the AP ligand. Applying SiMPull-POP to EphA2, a receptor tyrosine kinase, we found it to be mostly monomeric in the absence of its ligand, Ephrin-A1, and shifting toward higher-order oligomers upon ligand binding. To explore factors influencing ligand-independent assembly, we modulated membrane cholesterol content. Reducing cholesterol induced spontaneous EphA2 oligomerization, indicating that cholesterol suppresses receptor self-association. Overall, SiMPull-POP offers significant advantages over conventional techniques by enabling quantitative, single-molecule resolution of membrane protein complexes in a native-like environment. This approach provides critical insights into how membrane properties and external stimuli regulate protein assembly, supporting broader efforts to understand membrane protein function in both normal and disease states.
Key features
• Precise determination of membrane protein stoichiometry (e.g., monomer, dimer, oligomer) by directly counting photobleaching steps, overcoming the averaging limitations of bulk assays.
• By incorporating membrane proteins into DIBMA lipid particles (DIBMALPs), this preserves native lipid interactions, offering a more physiologically relevant context for studying protein assembly.
• Sensitively detects ligand-induced or membrane property-driven changes in oligomerization, making it a powerful tool for investigating both constitutive and regulated protein interactions.
Keywords: Single-molecule (单分子)Background
Experimentally capturing the complexity and dynamic nature of protein–protein or protein–lipid interactions in a native cellular environment remains a challenge [1,2]. Commonly used in situ methods to examine protein–protein interactions (i.e., resonance energy transfer imaging, two-hybrid screens, and co-immunoprecipitation) cannot easily preserve in vivo protein interactions due to the many steps between sample preparation and measurement and are blind to other heterogeneous interactions present in the sample [3]. Moreover, most techniques provide an ensemble measurement of changes in protein assembly rather than a distinct quantification of its oligomeric state. In order to quantitatively explore protein–protein interactions and the influence of lipids on these interactions in a more native context, we optimized and adapted a single-molecule pull-down (SiMPull) protocol previously reported [3,4]. Our advancement of the technique allows for retention of the native membrane environment by incorporating the co-polymer di-isobutylene maleic acid (DIBMA), which spawned the additional name SiMPull-POP (polymeric nanodisc photobleaching). SiMPull-POP is a method by which we can isolate proteins of interest directly from cell lysates and analyze them by total internal reflection fluorescence (TIRF) microscopy at single-molecule resolution (Figure 1).
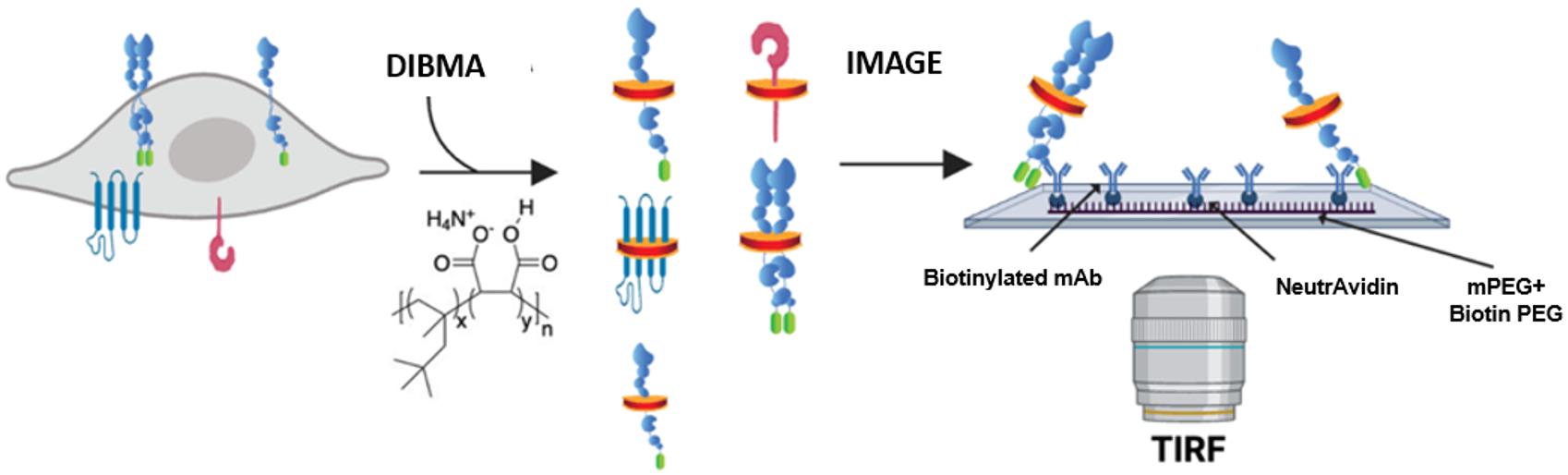
Figure 1. Schematic of single-molecule pull-down polymeric nanodisc photobleaching (SiMPull-POP). Cellular expression of a fluorescently tagged protein of interest, shown here as the GFP-tagged construct, that is extracted via membrane fractionation and solubilized with diisobutylene maleic acid (DIBMA). The solubilized sample is then flowed onto a Biotin-PEG/NeutrAvidin surface, incubated with a biotinylated antibody (“bait”) for the protein of interest (“prey”), and imaged by total internal reflection fluorescence (TIRF) microscopy. Figure modified from Schuck et al. [5].
The sensitivity of TIRF microscopy alleviates the need for large amounts of protein sample, as samples of interest can be visualized in the pM–nM range. In brief, SiMPull-POP begins by capturing diisobutylene maleic acid lipid particles (DIBMALPs) containing protein(s) of interest using a “bait and prey” type scenario constructed in a flow chamber using a microscope slide and a coverslip. In order to create such a capture system, the slide is passivated in a mixture of methoxy polyethylene glycol (mPEG) and biotinylated PEG (biotin-PEG) to prevent nonspecific adsorption of cell lysates and antibodies [6]. The surface is coated with NeutrAvidin, which interacts with biotin-PEG, followed by a biotinylated antibody against the protein of interest. As cell membrane extracts are passed through the flow chamber, the surface-bound antibody captures the protein of interest, while wash steps eliminate any unbound proteins, eliminating the need for laborious purification steps in a time-efficient manner. The sample is then subject to TIRF microscopy, where proteins of interest, bound to the slide surface, are visualized through excitation of a genetically encoded fluorescent protein tag on the protein of interest. By analyzing the fluorescence of individual molecules on the slide, over time, we can count individual photobleaching steps to determine how the protein of interest interacts with other proteins with the same tag. By binning photobleaching behaviors based on the number of steps observed, we are able to obtain a precise quantification of the membrane protein assembly state and begin to characterize factors regulating the assembly process.
Materials and reagents
Biological materials
1. Plasmid: pCMV6-EphA2-GFP (Origene, identifier: RG205725)
2. HEK293T cells (ATCC, CRL-3216)
3. Rabbit anti-EphA2 (D4A2) XP biotinylated (Cell Signaling Technologies, 97535), 20 nM of mAb stock in T50 buffer
4. Goat anti-GFP biotin conjugated (Rockland Immunochemical Inc., 600-106-215), 20 nM of mAb stock in T50 buffer
Reagents
1. DMEM (Thermo Fisher Scientific, catalog number: 11965092, 500 mL)
2. FBS (Thermo Fisher Scientific, catalog number: A5256701, 500 mL)
3. PenStrep (Thermo Fisher Scientific, catalog number: 15140122, 100 mL)
4. Opti-MEM reduced serum medium [Thermo Fisher Scientific, catalog number: 31985070 (500 mL) or 31985062 (100 mL)]
5. Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, catalog number: 11668030); see https://www.thermofisher.com/us/en/home/references/protocols/cell-culture/transfection-protocol/lipofectamine-2000.html for the manufacturer’s recommended reaction volumes and steps
6. Phosphate-buffered saline (PBS) (Thermo Fisher Scientific, catalog number: 10010023, 10× solution diluted to 1×)
7. Protease inhibitors (Fisher Scientific, catalog number: PIA32953)
8. Phosphatase inhibitors (Thermo Fisher Scientific, catalog number: A32957)
9. Diisobutylene maleic acid co-polymer (DIBMA) (Anatrace, catalog number: BMA101, 10% w/v in 1× PBS)
10. Pierce recombinant GFP (rGFP) (Thermo Fisher Scientific, catalog number: 88899, 100 nM stock)
11. NeutrAvidin protein (Thermo Fisher Scientific, catalog number: PI31000, 2 mg/mL prepared stock diluted to 0.2 mg/mL working stock)
12. Protocatechuic acid (PCA)/3,4-dihydroxybenzoic acid (Sigma-Aldrich, catalog number: 37580-25G-F, 100 mM stock in Milli-Q water, pH 9.0)
13. Bacterial protocatechuate 3,4-dioxygenase (rPCO), 300 units (Oriental Yeast Co., catalog number: 46852004)
14. Trolox (Acros Organics, catalog number: 2189400500)
Solutions
1. T50 buffer (see Recipes)
2. Lysis buffer (see Recipes)
3. Resuspension buffer (see Recipes)
4. Oxygen scavenging system (OSS) (see Recipes)
5. Trolox solution (see Recipes)
Recipes
1. T50 buffer
10 mM Tris-HCl
50 mM NaCl
pH = 8.0
2. Lysis buffer
50 mM Tris-HCl
250 mM sucrose
250 μM CaCl2
1 phosphatase inhibitor tablet (per 10 mL of lysis buffer)
1 protease inhibitor tablet (per 10 mL of lysis buffer)
pH = 7.4
3. Resuspension buffer
50 mM Tris-HCl
250 mM NaCl
Glycerol 90/10 (v/v)
pH = 8.0
4. Oxygen scavenging system (OSS)
1 mL of Trolox solution
40 μL of PCA
1 μL rPCO
Prepare fresh on the day of the experiment.
5. Trolox solution
100 mg of Trolox powder
40 mL of dH2O (shake overnight at 4 °C in dark)
Filter with a 0.2 μm syringe filter and store at 4 °C covered with aluminum foil or protected from light. Can be prepared and used for ~2–3 weeks. The solution should be clear; do not use it if coloration occurs.
Equipment
1. Optima Max-XP ultracentrifuge (Beckman Coulter, catalog number: 393315)
2. TLA55 rotor (Beckman Coulter, catalog number: 366725)
3. Sorvall Legend Micro 21R Centrifuge (Thermo Scientific, catalog number: 75002446)
4. Customized TIRF setup using an Inverted IX73 microscope frame (Olympus, water-immersion)
5. EMCCD camera (Andor Technology)
6. TIRF stage (TIRF Labs)
7. 465 nm cable laser (TIRF Labs)
8. 1″ × 3″ × 1 mm thick quartz slides (G. Finkenbeiner Inc., https://finkenbeiner.com/quartzslides)
9. Rectangular 1–1/2 cover glass (Corning Inc., catalog number: 2980-245)
10. Double-sided tape (Scotch, 3 M)
11. Scalpel (Fisher Scientific, catalog number: 22-444-272)
12. General-purpose grease [Dow Corning, MSC: 31735228 (mscdirect.com)]
13. Kimwipes (11 × 21 cm) [Kimtech Science, catalog number: 34155 (x60)/34120 (x30)]
14. 1.5 mL Eppendorf tubes (Fisher Scientific, catalog number: 05-408-129)
15. Microfuge tube polypropylene 1.5 mL, 9.5 × 38 mm 500 count (for ultracentrifugation) (Beckman Coulter, catalog number: 357448)
16. Air-Tite 1 mL syringes (Fisher Scientific, catalog number: 14-817-119)
17. 25-gauge needles (Fisher Scientific, catalog number: 14-826-49)
18. 27-gauge needles (Fisher Scientific, catalog number: 14-821-13B)
19. Costar 96-well plate, black opaque (Thermo Fisher Scientific, catalog number: 137101)
20. Plate reader (BioTek, model: Cytation 5)
21. Corning CytoSmart Cell Counter (Corning, catalog number: 6749)
22. Evos FLoid cell imaging station (Invitrogen, Thermo Fisher Scientific, catalog number: 4471136)
23. Falcon standard tissue culture dishes, 58.1 cm2 (Fisher Scientific, catalog number: 08-772E)
24. CO2 incubator (Panasonic, catalog number: MCO-170A1CUV-PA)
25. 0.2 μm syringe filter (Fisher Scientific, catalog number: 13-1001-06)
Software and datasets
1. GraphPad Prism, GraphPad Software Inc., https://www.graphpad.com
2. Microsoft Excel, Microsoft, https://www.microsoft.com
3. BioTek Gen5 Software for Imaging & Microscopy, Agilent, https://www.agilent.com/
4. IDL software, NV5 GEOSPATIAL SOFTWARE, https://www.nv5geospatialsoftware.com/Products/IDL
5. IDL Raw-Data-Analysis, Ha-SingleMoleculeLab, https://github.com/Ha-SingleMoleculeLab/
6. Single-molecule software, Ha-SingleMoleculeLab, https://github.com/Ha-SingleMoleculeLab/
7. Calculation to convert steps into % n-mer, https://github.com/sgouralis/composition_estimator
MatLab, https://www.mathworks.com/help/install/ug/install-products-with-internet-connection.html
8. Single-molecule photobleaching step binning code, Stefanski et al. [7]. Adapted from the Ha Lab scripts, https://github.com/justmwest/single-molecule-photobleaching/tree/master/count_photobleaching_steps
9. Anaconda Navigator + Spyder software, Anaconda, Anaconda.com
Anaconda 2.6 (older versions can be used through the newest update)
Python 3.12.4 (oldest) to the newest release
IPython 8.25.0 to the newest release
Spyder 5.5.1 to the newest release. Required import packages for Spyder: glob, os, random, numpy as np, sys, matplotlib. pyplot as plt, pandas as pd, from datetime import datetime, from tabulate import tabulate
Procedure
文章信息
稿件历史记录
提交日期: Aug 29, 2025
接收日期: Nov 28, 2025
在线发布日期: Dec 11, 2025
出版日期: Jan 5, 2026
版权信息
© 2026 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Schuck, R. J., Ward, A. E., Barrera, F. N. and Lamichhane, R. (2026). SiMPull-POP: Quantification of Membrane Protein Assembly via Single Molecule Photobleaching. Bio-protocol 16(1): e5560. DOI: 10.21769/BioProtoc.5560.
分类
生物物理学 > 单分子技术
生物化学 > 蛋白质 > 相互作用 > 蛋白质-蛋白质相互作用
生物化学 > 蛋白质 > 荧光
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link