- EN - English
- CN - 中文
Optimized Secretome Sample Preparation From High Volume Cell Culture Media for LC–MS/MS Proteomic Analysis
适用于 LC–MS/MS 蛋白质组学分析的大体积细胞培养上清分泌组样品制备方法优化
发布: 2025年12月20日第15卷第24期 DOI: 10.21769/BioProtoc.5542 浏览次数: 1096
评审: Ralph Thomas BoettcherPrajita PandeyAnonymous reviewer(s)
Abstract
The cellular secretome is a rich source of biomarkers and extracellular signaling molecules, but proteomic profiling remains challenging, especially when processing culture volumes greater than 5 mL. Low protein abundance, high serum contamination, and sample loss during preparation limit reproducibility and sensitivity in mass spectrometry–based workflows. Here, we present an optimized and scalable protocol that integrates (i) 50 kDa molecular weight cutoff ultrafiltration, (ii) spin column depletion of abundant serum proteins, and (iii) acetone/TCA precipitation for protein recovery. This workflow enables balanced recovery of both low- and high-molecular-weight proteins while reducing background from serum albumin, thereby improving sensitivity, reproducibility, and dynamic range for LC–MS/MS analysis. Validated in human mesenchymal stromal cell cultures, the protocol is broadly applicable across diverse cell types and experimental designs, making it well-suited for biomarker discovery and extracellular proteomics.
Key features
• Enables efficient concentration and cleanup of ≥5–500 mL of conditioned media, suitable for low-abundance secreted protein recovery.
• Combines 50 kDa ultrafiltration, optional HSA/IgG depletion, and acetone/TCA precipitation for robust removal of serum contaminants and improved signal-to-noise.
• Adaptable to various mammalian cell types and serum-free or serum-containing media; scalable for adherent and suspension cultures.
Keywords: Secretome (分泌组)Background
The secretome is a complete set of proteins secreted by cells into the extracellular space [1]. In a cell culture system, the secretome can be profiled from conditioned medium, which is a cell culture medium containing secreted proteins and signaling factors released by the cells during incubation under defined conditions [2]. Mass spectrometry–based proteomic analysis of the secretome is an established strategy for studying extracellular communication, discovering biomarkers, and identifying therapeutic targets [3,4]. In parallel, large-scale mapping of the human proteome highlights the importance of capturing both abundant housekeeping proteins and rare, tissue-specific factors in secretome studies [5]. However, secretome proteins are often present at very low concentrations and are masked by highly abundant serum proteins such as albumin and immunoglobulins [6], which complicates mass spectrometry analysis.
These challenges intensify when processing large volume samples (>5 mL), as often required to obtain sufficient secretome, particularly with primary cells or low-secreting cultures. Without rigorous sample processing, low-abundance proteins like cytokines and chemokines are frequently lost, while high-abundance proteins overshadow the results [6].
Early comparative work established in-gel digestion followed by LC–MS/MS as a robust strategy for protein identification, but this approach is labor-intensive, biased toward higher-abundance proteins, and suboptimal for low-molecular-weight cytokines [7]. More recently, systematic benchmarking studies demonstrated that acetone precipitation with in-solution digestion improved reproducibility and enhanced recovery of small-secreted proteins [8]. However, these protocols were primarily optimized for small input volumes (≤ 2 mL) and did not adequately address challenges of serum protein contamination or scalability.
To overcome these limitations, we developed an optimized protocol specifically designed for high-volume secretome samples (>5 mL) [9]. This workflow integrates three complementary steps: (i) 50 kDa ultrafiltration to generate both concentrate and filtrate fractions, (ii) spin column depletion [10] of abundant serum proteins, and (iii) acetone/TCA precipitation to maximize protein recovery. By processing both the filtrate and depleted concentrate in parallel, the protocol ensures balanced recovery of low-abundance cytokines and chemokines alongside high-molecular-weight extracellular matrix proteins, while minimizing background interference and serum protein contamination. The overall workflow is illustrated in Figure 1.
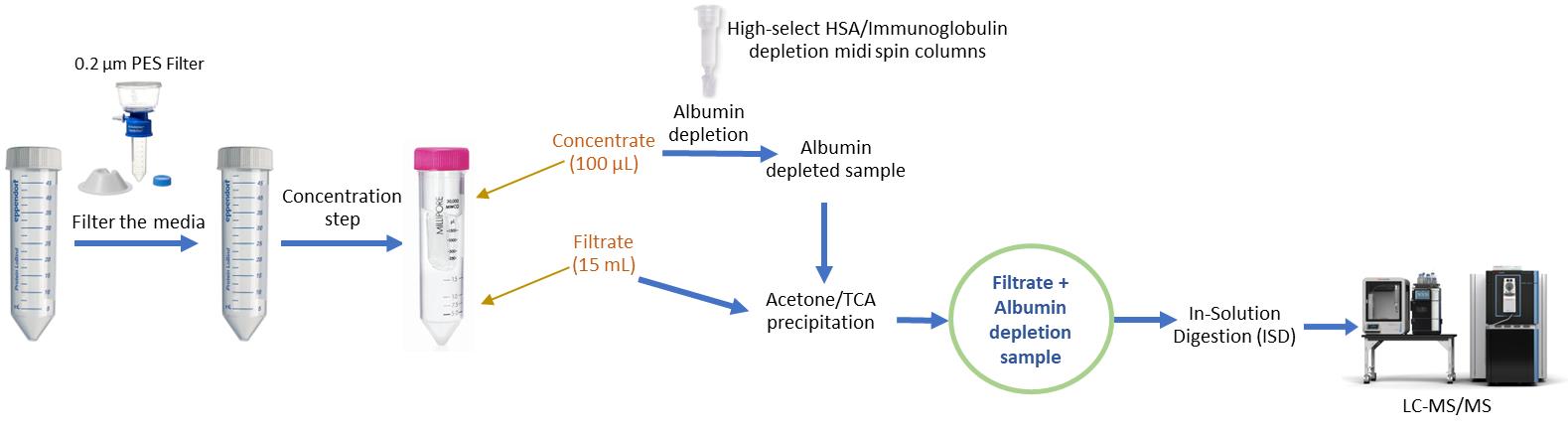
Figure 1. Workflow for optimized secretome preparation for LC–MS/MS analysis. Conditioned medium is first clarified by centrifugation (500× g, 5 min, 4 °C) and filtered through a 0.22 μm low-protein-binding filter to remove cells and debris. The clarified medium is then concentrated using Amicon Ultra-15 centrifugal filters (50 kDa MWCO), yielding approximately 100 μL of concentrate and 15 mL of filtrate, depending on the initial sample volume. The concentrate is subjected to HSA/IgG depletion to eliminate abundant serum proteins. The depleted concentrate and filtrate are then combined to generate a comprehensive secretome sample, which is subsequently precipitated, digested, and analyzed by LC–MS/MS. This workflow maximizes recovery of both low- and high-molecular-weight secreted proteins while minimizing background from serum components.
This method demonstrates improved sensitivity, reproducibility, and scalability compared to previous protocols. It maintains the identification capabilities of acetone precipitation workflows and extends their use to high-volume samples, which are often necessary for primary or low-secreting cultures. Additionally, it overcomes the reproducibility and coverage limitations associated with in-gel digestion-based approaches and outperforms ultrafiltration-only methods that do not efficiently recover low-molecular-weight proteins. Therefore, this optimized workflow offers a robust and adaptable solution for comprehensive secretome profiling, making it particularly advantageous for biomarker discovery pipelines that require broad dynamic range, scalability, and reproducibility.
Materials and reagents
Note: Catalog numbers and models are provided for reference; suitable alternatives may be used.
1. Amicon Ultra-15 centrifugal filter, 50 kDa MWCO (Merck Millipore, catalog number: UFC905008)
2. High-Select HSA/IgG depletion midi spin columns (Thermo Fisher Scientific, catalog number: A36367)
3. Chilled (-20 °C) acetone (Fisher Chemical, catalog number: A18-4)
4. Halt protease inhibitor cocktail (100×) (Thermo Scientific, catalog number: 1861278)
5. Trichloroacetic acid (TCA) (Fisher Chemical, catalog number: SA433-500)
6. Urea (Invitrogen, catalog number: 15505-035)
7. Ammonium bicarbonate (Fisher Chemical, catalog number: A643-500)
8. Dithiothreitol (DTT) (Fisher bioreagents, catalog number: BP172-5)
9. Iodoacetamide (IAA) (Sigma-Aldrich, catalog number: I1149 -5G)
10. MS-grade trypsin (Promega, catalog number: V5111)
11. Pierce C18 spin columns (Thermo Fisher Scientific, catalog number: 89870)
12. Formic acid (FA) (Thermo Scientific, catalog number: 28905)
13. Acetonitrile (ACN) (Fisher Chemical, catalog number: A955-4)
14. BCA Protein Assay kit (Thermo Fisher Scientific, catalog number: 23227)
15. Micro BCA Protein Assay kit (Thermo Fisher Scientific, catalog number: 23235)
16. Protein LoBind tube 2.0 mL (Eppendorf, catalog number: 022431102)
17. Centrifuge tube 50 mL flat cap (Thermo Fisher Scientific, catalog number: 05-539-13)
18. Nunc conical sterile polypropylene 15 mL centrifuge tubes (Thermo Scientific, catalog number: 339650)
19. Nalgene rapid-flow sterile disposable filter units with PES, CN, SFCA, or nylon membranes (Thermo Scientific, catalog number: 564-0020)
20. Mini-Protean TGX stain free gels (Bio-Rad, catalog number: 4568094)
21. Precision Plus protein dual color standards (Bio-Rad, catalog number: 1610374)
Equipment
1. Refrigerated benchtop centrifuge (Thermo Fisher Scientific, model: Sorvall X1R Pro-MD, Eppendorf, model: AG 5424)
2. SpeedVac concentrator (Thermo Scientific, model: Savant SPD111V, RVT5105)
3. NanoLC system (Thermo Scientific, model: Vanquish Neo)
4. Orbitrap mass spectrometer (Thermo Scientific, model: Orbitrap Exploris 240/480)
5. BCA reader (BioTek, model: Synergy/LX multi-mode reader)
6. Thermomixer (Eppendorf, model: Thermomixer C)
Procedure
文章信息
稿件历史记录
提交日期: Sep 16, 2025
接收日期: Nov 3, 2025
在线发布日期: Nov 20, 2025
出版日期: Dec 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Baby Mattamana, B., Gajjela, R., K.C., J., Parish, R. A. and Faull, P. A. (2025). Optimized Secretome Sample Preparation From High Volume Cell Culture Media for LC–MS/MS Proteomic Analysis. Bio-protocol 15(24): e5542. DOI: 10.21769/BioProtoc.5542.
分类
系统生物学 > 蛋白质组学 > 分泌蛋白质组
生物化学 > 蛋白质 > 分离和纯化
生物科学 > 生物技术 > 质谱
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link