- EN - English
- CN - 中文
Whole-Mount Visualization of Primary Cilia in the Developing Mouse Brain
发育中小鼠脑内初级纤毛的整体样本可视化分析
发布: 2025年12月20日第15卷第24期 DOI: 10.21769/BioProtoc.5538 浏览次数: 929
评审: Anonymous reviewer(s)

相关实验方案
玻璃体腔内NHS-生物素注射结合免疫组织化学标记与成像分析小鼠视神经中的蛋白运输
Caroline R. McKeown [...] Hollis T. Cline
2025年08月20日 2650 阅读
Abstract
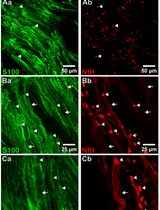
Primary cilia are evolutionarily conserved organelles that play critical roles in brain development. In the developing cortex, neural progenitors extend their primary cilia into the ventricular surface, where the cilia act as key signaling hubs. However, visualizing these cilia in a systematic and intact manner has been challenging. The commonly used cryostat sectioning only provides a limited snapshot of cilia on individual sections, and this process often disrupts the ciliary morphology. By contrast, the previously established whole-mount technique has been shown to preserve ciliary architecture in the adult mouse cortex. Here, we adapt and optimize the whole-mount approach for embryonic and neonatal brain, allowing robust visualization of ciliary morphology at the ventricular surface during development. This protocol describes step-by-step procedures for whole-mounting and immunostaining delicate embryonic and neonatal mouse cortices, enabling direct visualization of cilia in neural progenitors in the developing brain.
Key features
• This protocol adapts the whole-mount technique and applies it to delicate embryonic samples from embryonic day 12 (E12) to neonatal brain (P3).
• This protocol details the necessary steps to achieve intact and direct visualization of cilia in the developing mouse cortex.
• This protocol also provides the necessary steps for the dissection and visualization of cilia on the lateral ganglionic eminences (LGE) and medial ganglionic eminences (MGE).
Keywords: Primary cilia (初级纤毛)Background
Primary cilia are evolutionarily conserved microtubule-based organelles that project from the surface of most vertebrate cells and serve as signaling hubs. The primary cilium senses and transduces environmental cues into intracellular signaling pathways. Certain specialized cell types bear motile cilia, typically in multiple copies. Unlike primary cilia, motile cilia generate a rhythmical beating that propels the extracellular fluid or enables cellular motility [1–4]. Dysfunction of cilia leads to a wide range of diseases, collectively known as ciliopathies. Many ciliopathies, such as Joubert syndrome, Orofaciodigital syndrome, and Acrocallosal syndrome, are characterized by brain structural deficits [5,6]. These brain malformations often result in cognitive deficits and intellectual disabilities [3,5,7]. These deficits highlight the essential roles of primary cilia in normal brain development and neural circuitry formation.
During embryonic brain development, neurogenesis is precisely coordinated by radial glia (RG), the main neural progenitors in the developing brain. RG undergo various modes of cell division, producing the diverse neuronal cell types necessary to form functional neural circuits [8–13]. A notable anatomical feature of RG is that their radial processes extend across the entire developing cortex, from the pial (basal) surface to the ventricular (apical) surface [14]. While their nuclei undergo interkinetic nuclear migration—moving back and forth between the apical and basal sides in sync with the cell cycle—their centrosomes and primary cilia remain at the apical surface. In other words, the primary cilia of RG project into the cerebrospinal fluid (CSF) within the ventricles of the brain [15–17]. Studying the morphology of these primary cilia has been challenging. Conventional approaches, such as cryostat sectioning, are not ideal because they may physically damage the delicate structure of primary cilia. The freezing process itself, if not done properly, can cause the formation of ice crystals that distort the cilium. Furthermore, the mechanical slicing of the frozen tissue may shear or break the cilium, leading to artifacts that misrepresent its true morphology. Subsequent steps, such as mounting on slides, may further alter the structure. These damaged and distorted cilia make it difficult to get an accurate observation of their morphology. Finally, because primary cilia are thin, hair-like structures that project from the cell surface, a single section only captures a small, two-dimensional sliver of the cilium's full length. This fragmented view makes it nearly impossible to understand the cilium's overall shape, length, and orientation in its native three-dimensional context [18–22].
In this protocol, we adapt a whole-mount cortical preparation originally developed for the adult brain [23] to embryonic and neonatal brains. We aim to preserve intact ciliary morphology in their three-dimensional perspective at the native ventricular surface. By preserving the complete cytoarchitecture, this method enables a detailed study of ciliary morphology across different brain regions and at various developmental stages.
Materials and reagents
Biological materials
1. C57BL/6 (Jackson Laboratories, strain #000664) aged 6–8 weeks
Note: Embryonic (E) samples can be collected at E12, 14, and 17, and postnatal day (P) 1, 3, 5, and 7. The procedure is applicable to other mouse strains.
Reagents
1. DPBS (Fisher Scientific, catalog number: 14-190-136)
2. Triton X-100 (Fisher Scientific, catalog number: AAA16046AP)
3. Donkey serum (MilliporeSigma, catalog number: S30-100ML)
4. 20% Paraformaldehyde (PFA) (Fisher Scientific, catalog number: 50-980-493)
5. Hoechst (Sigma, catalog number: 94403-1ML)
6. Fluoromount-G mounting medium (SouthernBiotech, catalog number: 0100-01)
7. Rabbit anti-Arl13b (1:200) (Proteintech, catalog number: 17711-1-AP)
8. Rat anti-Arl13b (1:200) (BiCell Scientific, catalog number: 90413)
9. Mouse anti-Arl13b (1:200) (antibodiesinc, catalog number: 75-287
10. Mouse anti-acetylated tubulin (1:200) (Sigma, catalog number: T6793)
11. Mouse anti-β-catenin (1:100) (BD Biosciences, catalog number: 610154)
12. Rabbit anti-ZO1 (1:100) (Zymed, catalog number: 40-2200)
13. Mouse anti-AlexaFluor 488 (Jackson ImmunoResearch Labs, catalog number: 715-545-151)
14. Mouse anti-AlexaFluor rhodamine (Jackson ImmunoResearch Labs, catalog number: 715-025-151)
15. Mouse anti-AlexaFluor 647 (Jackson ImmunoResearch Labs, catalog number: 715-605-151)
16. Rabbit anti-AlexaFluor 488 (Jackson ImmunoResearch Labs, catalog number: 711-545-152)
17. Rabbit anti-AlexaFluor rhodamine (Jackson ImmunoResearch Labs, catalog number: 711-025-152)
18. Rabbit anti-AlexaFluor 647 (Jackson ImmunoResearch Labs, catalog number: 711-605-152)
19. Rat anti-AlexaFluor 488 (Jackson ImmunoResearch Labs, catalog number: 712-545-153)
20. Rat anti-AlexaFluor rhodamine (Jackson ImmunoResearch Labs, catalog number: 712-025-153)
21. Rat anti-AlexaFluor 647 (Jackson ImmunoResearch Labs, catalog number: 712-605-153)
Solutions
1. Blocking buffer (see Recipes)
Recipes
1. Blocking buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Donkey serum | 2% | 2 mL |
| Triton X-100 | 0.2% | 200 μL |
| DPBS | n/a | 97.8 mL |
| Total | n/a | 100 mL |
Store at 4 °C.
Laboratory supplies
1. FisherbrandTM surface treated tissue culture dishes, 10 cm dish (Fisher Scientific, catalog number: FB012924)
2. Thermo ScientificTM BioLite 6 cm cell culture treated dishes (Fisher Scientific, catalog number: 12-556-001)
3. 96-well plate (Thermo Fisher Scientific, catalog number: 12-556-008)
4. 24-well plate (Fisher Scientific, catalog: FB012929)
5. AxygenTM MaxyClear Snaplock microtubes, 1.5 mL (Axygen, catalog number: 14-222-155)
6. CorningTM 15 mL centrifuge tubes with CentriStarTM cap (Fisher Scientific, catalog number: 430791)
7. FisherbrandTM SuperfrostTM Plus microscope slides (Fisher Scientific, catalog number: 12-550-15)
8. FisherbrandTM SuperslipTM coverslips (Fisher Scientific, catalog number: 12-541-056)
9. Moria ultra-fine forceps (Fine Science Tools, catalog number: 11370-40)
10. Moria ultra-fine curved forceps (Fine Science Tools, catalog number: 11370-42)
11. Extra fine Bonn 8.5 cm scissors (Fine Science Tools, catalog number: 14085-08)
Equipment
1. Confocal microscope (Zeiss, model: LSM 880)
2. Leica Mica
Note: All other fluorescence microscopes can also be used with 63× lens or above.
3. Vortex mixer (Fisher Scientific, catalog number: 02-215-414)
4. Centrifuge (Eppendorf, model: 5702R)
Software and datasets
1. LAS X (Leica, v3.7.4.23463)
2. Fiji (Version 1.54p)
Procedure
文章信息
稿件历史记录
提交日期: Sep 23, 2025
接收日期: Nov 2, 2025
在线发布日期: Nov 19, 2025
出版日期: Dec 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Gutierrez, O. T., Liu, X. and Ge, X. (2025). Whole-Mount Visualization of Primary Cilia in the Developing Mouse Brain. Bio-protocol 15(24): e5538. DOI: 10.21769/BioProtoc.5538.
分类
发育生物学 > 器官形成 > 大脑
细胞生物学 > 细胞结构 > 细胞表面
神经科学 > 神经解剖学和神经环路 > 免疫荧光
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link