- EN - English
- CN - 中文
Quantitative Analysis of the Arabidopsis Leaf Secretory Proteome via TMT-Based Mass Spectrometry
基于 TMT 的质谱技术对拟南芥叶片分泌蛋白组的定量分析
(*contributed equally to this work) 发布: 2025年11月20日第15卷第22期 DOI: 10.21769/BioProtoc.5508 浏览次数: 2016
评审: Neha NandwaniFernando A Gonzales-ZubiateAnonymous reviewer(s)

相关实验方案

通过简并PCR鉴定二倍体马铃薯Solanum okadae中的S位点F-box蛋白序列
Amar Hundare [...] Timothy P. Robbins
2025年06月05日 1991 阅读
Abstract
In plants, the apoplast contains a diverse set of proteins that underpin mechanisms for maintaining cell homeostasis, cell wall remodeling, cell signaling, and pathogen defense. Apoplast protein composition is highly regulated, primarily through the control of secretory traffic in response to endogenous and environmental factors. Dynamic changes in apoplast proteome facilitate plant survival in a changing climate. Even so, the apoplast proteome profiles in plants remain poorly characterized due to technological limitations. Recent progress in quantitative proteomics has significantly advanced the resolution of proteomic profiling in mammalian systems and has the potential for application in plant systems. In this protocol, we provide a detailed and efficient protocol for tandem mass tag (TMT)-based quantitative analysis of Arabidopsis thaliana secretory proteome to resolve dynamic changes in leaf apoplast proteome profiles. The protocol employs apoplast flush collection followed by protein cleaning using filter-aided sample preparation (FASP), protein digestion, TMT-labeling of peptides, and mass spectrometry (MS) analysis. Subsequent data analysis for peptide detection and quantification uses Proteome Discoverer software (PD) 3.0. Additionally, we have incorporated in silico–generated spectral libraries using PD 3.0, which enables rapid and efficient analysis of proteomic data. Our optimized protocol offers a robust framework for quantitative secretory proteomic analysis in plants, with potential applications in functional proteomics and the study of trafficking systems that impact plant growth, survival, and health.
Key features
• Rapid and high-purity collection of Arabidopsis thaliana leaf apoplast flush.
• Use of filter-aided sample preparation (FASP) for protein cleaning to obtain high-quality data.
• Use of in-house-generated theoretical spectral libraries for efficient and rapid analysis of MS data.
Keywords: Arabidopsis (拟南芥)Graphical overview
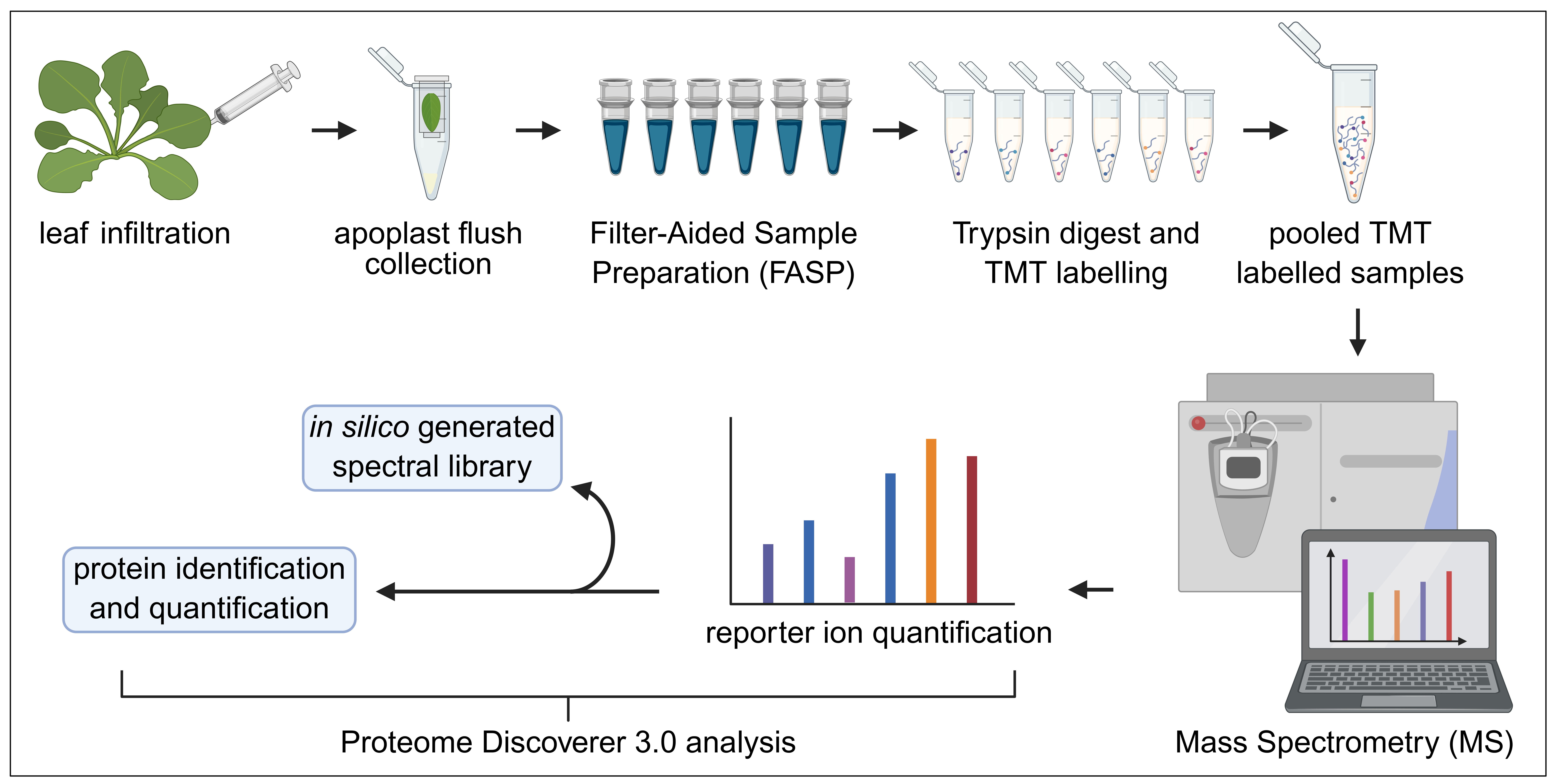
Tandem mass tag (TMT)-mass spectrometry (MS) analysis of Arabidopsis secretory proteome
Background
Plants, as sessile organisms, are constantly exposed to multifactorial abiotic and biotic stresses in both natural and agricultural environments. The apoplast serves as a critical interface between the plant cell and its environment, containing a dynamic array of secretory proteins that contribute to cell wall remodeling, cell signaling, abiotic stress, and host-defense mechanisms [1–3]. In order to survive in a challenging environment, plants must dynamically regulate apoplast protein composition, relying on a combination of constitutive and environment-sensitive secretory pathways active at the cell membrane [4–8]. To understand plant adaptations to climate change and elucidate secretory mechanisms in plant cells, a detailed understanding of dynamic changes in the plant secretory proteome profile is crucial. The knowledge gained will feed future strategies and interventions to enhance plant immunity and resilience to climate change, thereby improving agricultural production.
Plant proteomics research is gaining momentum for the high-throughput profiling of secretory proteins [1]. This process typically involves liquid chromatography (LC) coupled to tandem mass spectrometry (MS/MS) and bioinformatics technology. A label-free quantification technique is commonly utilized for apoplast profiling. Although the label-free technique offers benefits such as cost-effectiveness, higher proteome coverage, and flexibility, its major disadvantage is lower accuracy, sensitivity, and limited multiplexing [9–11]. In contrast, label-based quantification, such as tandem mass tag (TMT), offers high precision and accuracy, facilitating multiplexing up to 35 samples. TMT kits are commercially available in 6-, 10-, 11-, 16-, 18-, and 35-plex formats, allowing user selection based on their specific experimental needs [12–14] and enabling simultaneous quantitative analysis of samples under different conditions and biological replicates in a single run [15–17]. TMT-labeled spectral libraries, collections of reference spectra representing peptides, are commonly used in other eukaryotes and prokaryotes, yet they remain rare in plant proteomics. We outline here a protocol that harnesses this TMT labeling strategy for quantitative plant proteome profiling.
Plant secretory proteomic analysis is a complex multi-step experimental workflow that relies on efficient sample collection, combined with biochemistry, proteomics, and bioinformatics in a precise manner, with each step posing distinct challenges. Common challenges include cytoplasmic protein and nonprotein contaminants, which can affect sample processing in subsequent stages as well as proteomic analysis [18]. Highly abundant cytoplasmic contaminants, such as RUBISCO, hinder MS analysis by overshadowing the low-abundant secretory proteins. During apoplast flush collection, cytoplasmic leakage due to cell damage cannot be completely avoided. Therefore, it is important that sample preparation is optimized to minimize cytosolic contamination and ensure uniformity in sample preparation for reliable data collection.
There are several advanced tools available for MS data analysis, but their use in plants is limited. Compared to conventional sequence searching, spectral library searching is faster and provides higher specificity and sensitivity [19]. While unlabeled spectral libraries for Arabidopsis’s proteome are available [20], the TMT-labeled spectral libraries are lacking. Proteome Discoverer 3.0/3.1 (PD 3.0/3.1) allows the prediction of TMT-labeled spectral libraries, effectively addressing this gap. We successfully utilized TMT-based (TMT 6-plex, 11-plex, and 18-plex) quantitative mass spectrometry to characterize the Arabidopsis secretory proteomic profiles from the leaf apoplast [21]. In doing so, we were able to demonstrate for the first time that TMT–MS analysis, coupled with analysis using predicted spectral libraries via Proteome DiscovererTM software, achieves enhanced peptide detection. The strategy has enabled high-resolution proteome analysis to resolve the mechanism of the SNARE-assisted secretory pathways during bacterial pathogenesis in plants.
This protocol focuses on the analysis of the Arabidopsis secretory proteome using the TMT 6-plex approach. The protocol consists of the following major steps: apoplast flush collection, protein cleaning, digestion, TMT labeling, MS analysis, and subsequent data analysis using PD3.0 (see Figure 1). The primary benefits of this optimized protocol include (1) rapid collection of apoplast flush in bulk amount with minimal cytoplasmic protein contamination, (2) filter-aided sample preparation (FASP) for protein cleaning, consistently generating high-quality data, (3) the use of spectral libraries for efficient and rapid analysis of MS data, and (4) scalable method up to 10-, 11-, 16-, and 18-plex TMT labeling and analysis. For further details, including the results and analysis process, please refer to Waghmare et al. [21].

Figure 1. Workflow diagram
Materials and reagents
CAUTION: General laboratory safety precautions should be taken while working with flammable, corrosive, and toxic chemicals, including the use of a fume hood and appropriate personal protective equipment, while adhering to regulations that follow standard health and safety risk assessments.
Note: Store reagents as per the manufacturer’s instructions and check use-by date.
Biological materials
1. Arabidopsis thaliana Columbia-0 ecotype (Col-0, ABRC stock CS60000) and transgenic lines; store seeds at 4 °C in dry conditions for maximum viability
Reagents
1. Sodium dodecyl sulphate (SDS) (Sigma-Aldrich, catalog number: L4509)
2. Tris base (Sigma-Aldrich, catalog number: T6066)
3. Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D9163)
4. Urea (Sigma-Aldrich, catalog number: U5128)
5. HCl (Fisher Chemical, catalog number: 10294190)
6. Iodoacetamide (IAA) (Sigma-Aldrich, catalog number: 1149)
7. Ammonium bicarbonate (Sigma-Aldrich, catalog number: A6141)
10. Trypsin (Promega, catalog number: V5111)
12. Acetonitrile (Rathburn LCMS-Grade, catalog number: RH1016LCMS)
13. Triethylammonium bicarbonate (TEAB) (Sigma-Aldrich, catalog number: T7408)
14. MgCl2 (Sigma-Aldrich, catalog number: M8266)
15. Laemmli buffer (Bio-Rad, catalog number: 1610747)
16. Gel stain (Neo Biotech, catalog number: NB-45-00078)
17. Bradford protein assay dye reagent concentrate (Bio-Rad, catalog number: 5000006)
18. Acetonitrile (HPLC-grade) (Rathburn LCMS-Grade, catalog number: RH1016LCMS)
19. Formic acid (Thermo Fisher Scientific, catalog number: A117)
20. TFA (Thermo Fisher Scientific, catalog number: 85183)
21. TMT Isobaric Mass Tagging Kit, 6-plex, 10-plex, 16-plex (Thermo Fisher Scientific)
22. Hydroxylamine (Thermo Fisher Scientific, catalog number: 90115)
23. Distilled water
Solutions
1. Infiltration buffer (see Recipes)
2. SDT-lysis buffer (see Recipes)
3. 0.1 M Tris/HCl (see Recipes)
4. UA (see Recipes)
5. IAA (see Recipes)
6. Trypsin (see Recipes)
7. 10% acetonitrile (see Recipes)
8. 50 mM ammonium bicarbonate (see Recipes)
9. 100 mM TEAB (see recipes)
10. 5% hydroxylamine (quenching solution) (see Recipes)
11. Reverse-phase UHPLC solvent A (see Recipes)
12. Reverse-phase UHPLC solvent B (see Recipes)
13. Sample resuspension buffer (see Recipes)
Recipes
1. Infiltration buffer
| Reagent | Final concentration | Quantity for 100 mL |
|---|---|---|
| 1 M MgCl2 | 10 mM | 1 mL |
| Autoclaved MilliQ water | 99 mL |
Note: Store at 4 °C.
2. SDT-lysis buffer
| Reagent | Final concentration | Quantity for 100 mL |
| 10% SDS | 4% (w/v) | 40 mL |
| 1 M Tris/HCl (pH 7.6) | 100 mM | 10 mL |
| 1 M DTT | 100 mM | 10 mL |
| Autoclaved MilliQ water | to 100 mL |
Note: Store at room temperature.
Caution: Wear a mask and gloves when handling SDS powder to avoid dust inhalation.
3. 0.1 M Tris/HCl
| Reagent | Final concentration | Quantity for 1 L |
| 1 M Tris/HCl (pH 8.5) | 0.1 M | 100 mL |
| Autoclaved MilliQ water | 900 mL |
Note: Store at 4 °C.
4. UA
| Reagent | Final concentration | Quantity for 10 mL |
| 8.8 M Urea | 8 M | 9 |
| 1 M Tris/HCl (pH 8.5) | 100 mM | 1 |
Note: Prepare freshly and use within 1 day. 1 mL per sample.
5. IAA
| Reagent | Final concentration | Quantity for 1 mL |
| 0.5 M iodoacetamide | 50 mM | 100 μL |
| UA | 900 μL |
Note: Store at 4 °C. Prepare freshly and use within 1 day. 0.1 mL per sample.
6. Trypsin
| Reagent | Final concentration | Quantity for 100 μL |
| 1 μg/μL trypsin | 0.05 μg/μL | 5 μL |
| 1 M ammonium bicarbonate (pH 8.3) | 50 mM | 95 μL |
Note: Store at -20 °C. The enzyme-to-protein ratio should be 1:100. For 25 μg of protein, use 0.25 μg of trypsin.
7. 10% acetonitrile
| Reagent | Final concentration | Quantity for 1 L |
| Acetonitrile | 10% | 100 mL |
| Autoclaved MilliQ water | 900 mL |
Note: Store at room temperature.
8. 50 mM ammonium bicarbonate
| Reagent | Final concentration | Quantity for 100 mL |
| 1 M ammonium bicarbonate (pH 8.3) | 50 mM | 5 mL |
| Autoclaved MilliQ water | 95 mL |
Note: Prepare 0.5 mL per 1 sample. Store at -20 °C.
9. 100 mM TEAB
| Reagent | Final concentration | Quantity for 1 L |
| 1 M triethylammonium bicarbonate | 100 mM | 100 mL |
| Autoclaved MilliQ water | 900 mL |
Note: Store at -20 °C.
10. 5% hydroxylamine (quenching solution)
| Reagent | Final concentration | Quantity for 1 mL |
| 50% hydroxylamine | 5% | 0.1 mL |
| Autoclaved MilliQ water | 0.9 mL |
Note: Store at room temperature.
Caution: Wear a mask and gloves when handling. Avoid release to the environment.
11. Reverse-phase UHPLC solvent A
| Reagent | Final concentration | Quantity for 100 mL |
| Formic acid | 0.01% | 10 μL |
| Autoclaved MilliQ water | to 100 mL |
Note: Store at room temperature.
12. Reverse-phase UHPLC solvent B
| Reagent | Final concentration | Quantity for 100 mL |
| Formic acid | 0.08% | 80 μL |
| Acetonitrile | 80% | 80 mL |
| Autoclaved MilliQ water | 20% | to 100 mL |
Note: Store at room temperature.
13. Sample resuspension buffer
| Reagent | Concentration | Quantity for 10 mL |
| Formic acid | 0.50% | 50 μL |
| Acetonitrile | 1% | 100 μL |
| Autoclaved MilliQ water | to 10 mL |
Note: Store at room temperature.
Laboratory supplies
1. Soil mix
2. Plant pots (with holes)
3. Plant trays without drain holes
4. Plant growth room/chamber
5. Forceps, sticky tapes, markers
6. Wash bottles for watering (500 mL)
7. Falcon tubes 15 mL (Corning, catalog number: 430790) and 50 mL (Corning, catalog number: 430828)
8. Microfuge tubes 1.5 mL (Sarstedt, catalog number: 72.690.001) and 0.5 mL (Sarstedt, catalog number: 72.699)
9. Syringes 1 mL (Thermo Fisher Scientific, catalog number: 15489199) and 10 mL (Thermo Fisher Scientific, catalog number: 15205093)
10. Disposable gloves, forceps, paper towels, blades
11. Micropipettes (100 μL, 1 mL)
12. Protein ladder (Thermo Fisher Scientific, catalog number: 26619)
13. NuPAGE 4%–12% bis-tris gel (Invitrogen, catalog numbers: NP0321BOX and NP0322BOX)
14. Filter unit (Millipore, model: Ultracel YM-30)
15. 96-well plate (Greiner Bio-One, catalog number: 651101)
16. C18 trap column (0.3 × 5 mm) PepMap Neo C18 5 μm, 300 μm × 5 mm 17400 (Thermo Scientific)
17. Acclaim Pepmap RSLC C18 reversed phase column (50 cm × 75 μm nano viper, particle size 2 μm, pore size 100 Å) (Thermo Scientific, model: SN20579764)
Equipment
1. Vortex (Scientific Industries, model: Vortex-Genie® 2 mixer)
2. Autoclave (Rodwell, model: ST2228V)
3. Refrigerated centrifuge (fixed angle rotor) (Eppendorf, model: 5424R, fixed-angle rotor)
4. Powerpack
5. SDS-PAGE gel apparatus (Life Technology, catalog number: A25977)
6. Wet chamber/heat block set to 37 °C
7. Liquid chromatography (LC): Dionex Ultimate 3000 RSLC nanoflow system (Dionex, Camberley, UK)
8. Electrospray ionization (ESI) mass spectrometry: Orbitrap Elite/fusion (Thermo Fisher Scientific, UK)
Software and datasets
1. Database: Arabidopsis taxonomy from the NCBI database
2. Software: Proteome Discoverer 3.0 software (Thermo Fisher Scientific, UK)
Procedure
文章信息
稿件历史记录
提交日期: Jul 22, 2025
接收日期: Oct 8, 2025
在线发布日期: Oct 21, 2025
出版日期: Nov 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Waghmare, S., Xia, L., McGill, S., Burchmore, R. and Karnik, R. (2025). Quantitative Analysis of the Arabidopsis Leaf Secretory Proteome via TMT-Based Mass Spectrometry. Bio-protocol 15(22): e5508. DOI: 10.21769/BioProtoc.5508.
- Waghmare, S., Xia, L., Ly, T. P., Xu, J., Farami, S., Burchmore, R., Blatt, M. R. and Karnik, R. (2025). SYNTAXIN OF PLANTS 132 underpins secretion of cargoes associated with salicylic acid signaling and pathogen defense. Plant Physiol. 197(1). e1093/plphys/kiae541. https://doi.org/10.1093/plphys/kiae541
分类
植物科学 > 植物生物化学 > 蛋白质
植物科学 > 植物分子生物学 > 蛋白质
系统生物学 > 蛋白质组学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link