- EN - English
- CN - 中文
A Practical CRISPR-Based Method for Rapid Genome Editing in Caulobacter crescentus
基于CRISPR的快速基因组编辑方法在新月弧菌中的应用
发布: 2025年11月05日第15卷第21期 DOI: 10.21769/BioProtoc.5490 浏览次数: 1807
评审: Alessandro DidonnaAnonymous reviewer(s)

相关实验方案
I-PREFR:基于反向PCR的无酶单向策略,利用自杀载体在细菌中快速实现无标记染色体基因缺失与重建
Rekha Rana [...] Prabhu B. Patil
2025年05月20日 2342 阅读
Abstract
The RNA-guided Cas enzyme specifically cuts chromosomes and introduces a targeted double-strand break, facilitating multiple kinds of genome editing, including gene deletion, insertion, and replacement. Caulobacter crescentus and its relatives, such as Agrobacterium fabrum and Sinorhizobium meliloti, have been widely studied for industrial, agricultural, and biomedical applications; however, their genetic manipulations are usually characterized as time-consuming and labor-intensive. C. crescentus and its relatives are known to be CRISPR/Cas-recalcitrant organisms due to intrinsic limitations of SpCas9 expression and possible CRISPR escapes. By fusing a reporting gene to the C terminus of SpCas9M and precisely manipulating the expression of SpCas9M, we developed a CRISPR/SpCas9M-reporting system and achieved efficient genome editing in C. crescentus and relatives. Here, we describe a protocol for rapid, marker-less, and convenient gene deletion by using the CRISPR/SpCas9M-reporting system in C. crescentus, as an example.
Key features
• CRISPR-SpCas9M-reporting system overcomes the limitation of CRISPR escape and achieves a high apparent editing efficiency.
• The method enables multiple kinds of genome editing, generating in-frame and marker-less chromosomal modifications.
• The method completes a cycle of genome editing within one week.
• The method could be readily applied for genome editing in C. crescentus, A. fabrum, and S. meliloti.
Keywords: CRISPR (CRISPR)Graphical overview
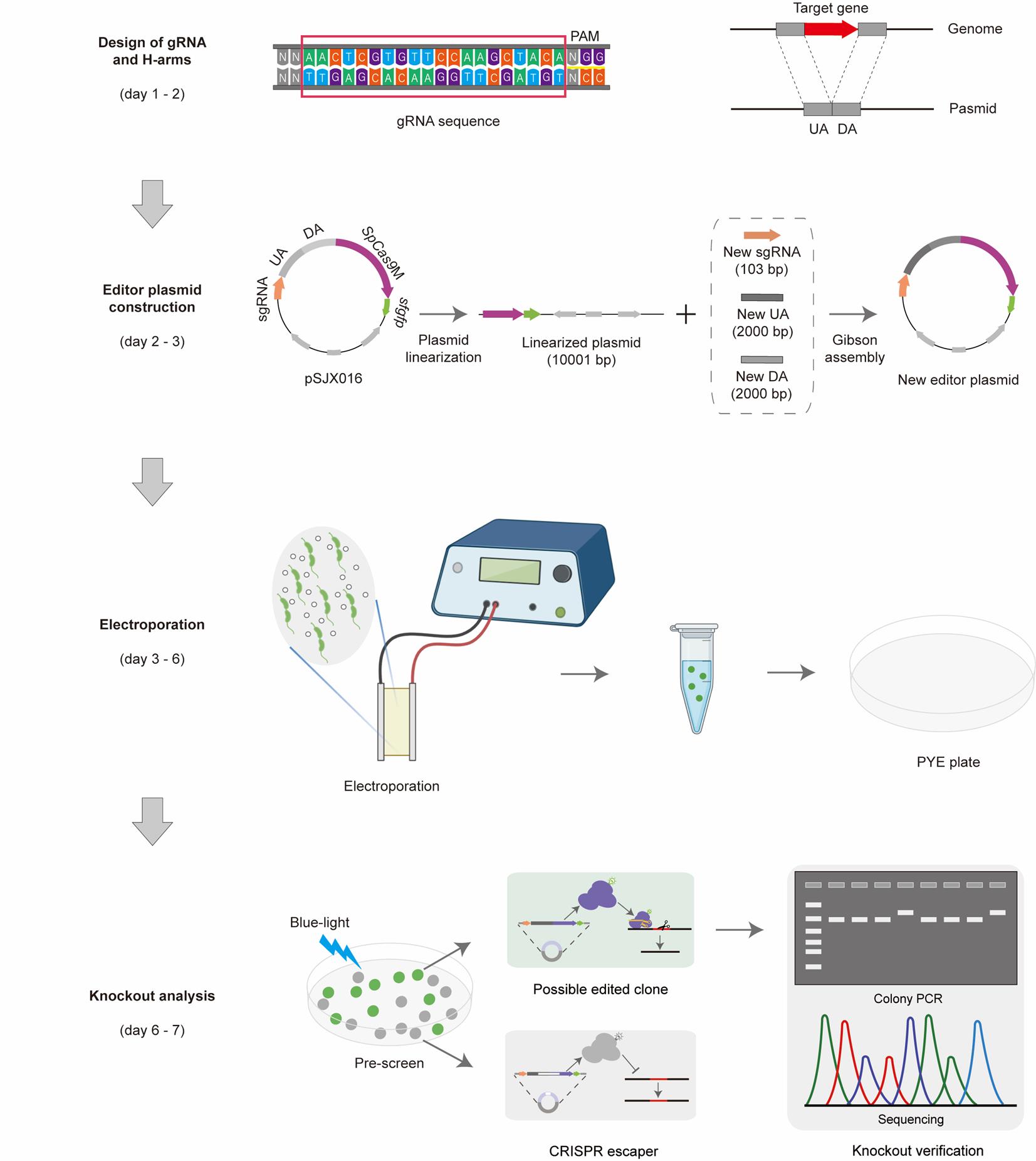
Workflow of gene deletion using the CRISPR/SpCas9M-reporting system. Step 1: Design of gRNA and H-arms. All the gRNA sequences were designed in CHOPCHOP [1]. H-arms consist of the upstream arm (UA, 2,000 bp) and the downstream arm (DA, 2,000 bp) of the targeted gene. Step 2: Editor plasmid construction. The starting plasmid pSJX016 was used as a starting point by replacing the sgRNA and H-arms in just one round of Gibson assembly. The corrected editor plasmids were checked by PCR and sequencing. Step 3: Electroporation. The new editor plasmid was transferred into the competent cells of C. crescentus through electroporation, and the possible edited cells were first selected on the PYE plates with the corresponding resistance. Step 4: Knockout analysis. The clones normally expressing SpCas9M were prescreened by the indication of fluorescent sfGFP under a blue-light lamp, and edited clones were finally confirmed by colony PCR and sequencing.
Background
Caulobacter crescentus, a member of Caulobacterales within α-proteobacteria, is a free-living bacterium widely distributed in freshwater environments [2,3]. Due to its tightly regulated cell cycle and asymmetric cell division, C. crescentus serves as an excellent model for studying cell polarity and cell differentiation [4–6]. The current genetic tool for chromosomal deletions and insertions in C. crescentus was first established in 1991 by Ely et al., utilizing sacB-based counterselection and allelic exchange via homologous recombination (HR) [7]. However, this method is labor-intensive and exhibits a relatively low editing efficiency [8]. In 2006, Martin Thanbichler and Lucy Shapiro et al. reported a single-crossover integration strategy that enables efficient gene insertion in C. crescentus [9,10]. Despite its greatly improved efficiency, this approach integrates the entire editing plasmid (including the selection marker) into the genome and is unsuitable for gene knockout.
CRISPR technology enables precise genome editing by utilizing a vector encoding the Cas enzyme and single-guide RNA (sgRNA). Guided by the sgRNA, the Cas enzyme induces a double-strand break (DSB) at the target sequence [11,12]. The corresponding cells then repair the DSB through either non-homologous end joining (NHEJ) or homology-directed repair (HDR), facilitating targeted gene knockout or knock-in [13]. Unlike the first-generation zinc finger nucleases (ZFNs) and the second-generation transcription activator-like effector nucleases (TALENs), CRISPR relies on the base pairing between gRNA and target DNA rather than protein–DNA binding interactions [14]. This enables CRISPR to exhibit distinct advantages, including both accuracy (editing specificity) and practicality (speed and cost-effectiveness).
For decades, scientists have attempted to develop efficient genome-editing tools for C. crescentus and its relatives via HR-based approaches. However, these efforts have largely been unsuccessful [15–17]. To address the challenges, we developed a CRISPR/Cas-assisted HR method for C. crescentus using an all-in-one editor plasmid, named CRISPR/SpCas9M-reporting system [18]. The reporting system facilitates the identification of CRISPR escape events and increases the apparent editing efficiency in C. crescentus. This method enables targeted gene deletion and insertion within less than a week, producing in-frame and marker-less edits on the chromosomes. Moreover, it has been successfully applied to two C. crescentus relatives, Agrobacterium fabrum and Sinorhizobium meliloti, establishing it as a general editing strategy [18]. We anticipate that this practical tool could be applicable to other difficult-to-edit organisms, facilitating both basic and applied research in α-proteobacteria.
Materials and reagents
Biological materials
1. C. crescentus NA1000 (Lucy Shapiro lab, Department of Developmental Biology, Stanford University School of Medicine)
2. Trans5α chemically competent cell (Transgen Biotech, catalog number: CD201-01)
3. pSJX016 plasmid (our lab, Shenzhen Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology)
Reagents
1. PCR primers (Sangon Biotech), standard 25–50 nmol scale
2. 2× Phanta Flash Master Mix (Vazyme, catalog number: P520-01/02/03)
3. KOD OneTM PCR Master Mix (TOYOBO, catalog number: 450800)
4. Agarose (Baygene Biotech, catalog number: 232475)
5. BactoTM peptone (BD, catalog number: 211677)
6. BactoTM yeast extract (BD, catalog number: 212750)
7. BactoTM agar (BD, catalog number: 214010)
8. Magnesium sulfate (MgSO4) (Sigma, catalog number: 10034-99-8)
9. SYBRTM Safe DNA gel stain (Invitrogen, catalog number: 2978645)
10. Ethanol absolute (Macklin, catalog number: 64-17-5)
11. 50× TAE (Sangon Biotech, catalog number: B548101-0500)
12. LB broth (Huankai Microbial, catalog number: 028324)
13. LB agar (Huankai Microbial, catalog number: 028334)
14. Gel Extraction kit (Omega, catalog number: D2500020000E20Y090)
15. EasyPure Plasmid MiniPrep kit (TransGen Biotech, catalog number: EM101-02)
16. ClonExpress MultiS One Step Cloning kit (Vazyme, catalog number: C113-01/02)
17. FastDigest DpnI (Thermo Scientific, catalog number: FD1704)
18. 10× FastDigest buffer (Thermo Scientific, catalog number: FD1704)
19. DNA marker (Accurate Biology, catalog number: AG11909)
20. Kanamycin (Sangon Biotech, catalog number: A600286-0025)
21. ddH2O
22. Vanillic acid (Sigma, catalog number: 34770-10G)
23. MgSO4 (Sigma, catalog number: 10043-52-4)
24. CaCl2 (Sigma, catalog number: 10034-99-8)
Solutions
1. Kanamycin stock solution (50 mg/mL) (see Recipes)
2. LB agar medium (see Recipes)
3. LB liquid medium (see Recipes)
4. Peptone yeast extract (PYE) agar medium (see Recipes)
5. PYE liquid medium (see Recipes)
Recipes
1. Kanamycin stock solution (50 mg/mL)
a. Dissolve 0.5 g of kanamycin (powder) in 10 mL of distilled water at room temperature and filter the solution through a 0.2 μm filter. Then, aliquot the solution into 1 mL fractions and store in a -20 °C freezer.
b. When necessary, supplement media with kanamycin at the following concentrations: 5/25 and 50/50 (liquid/solid media for C. crescentus and for E. coli, respectively, in μg/mL).
2. LB agar medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Bacto tryptone | 1% (w/v) | 10 g |
| Yeast extract | 0.5% (w/v) | 5 g |
| Agar | 1.5% (w/v) | 15 g |
| NaCl | 0.5% (w/v) | 5 g |
| H2O | n/a | Fill up to 1 L |
| Total | n/a | 1 L |
3. LB liquid medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Bacto tryptone | 1% (w/v) | 10 g |
| Yeast extract | 0.5% (w/v) | 5 g |
| NaCl | 0.5% (w/v) | 5 g |
| H2O | n/a | Fill up to 1 L |
| Total | n/a | 1 L |
4. PYE agar medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Peptone | 0.2% (w/v) | 2 g |
| Yeast extract | 0.1% (w/v) | 1 g |
| Bacto agar | 1.5% (w/v) | 15 g |
| 1 M MgSO4 | 1 mM | 1 mL |
| 1 M CaCl2 | 0.5 mM | 0.5 mL |
| H2O | n/a | Fill up to 1 L |
| Total | n/a | 1 L |
5. PYE liquid medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Peptone | 0.2% (w/v) | 2 g |
| Yeast extract | 0.1% (w/v) | 1 g |
| 1 M MgSO4 | 1 mM | 1 mL |
| 1 M CaCl2 | 0.5 mM | 0.5 mL |
| H2O | n/a | Fill up to 1 L |
| Total | n/a | 1 L |
Laboratory supplies
1. Petri dishes (Fisher Scientific, catalog number: FB0875712)
2. 0.22 μm Millipore filter (Millipore, catalog number: R7MA69670)
3. 0.2 mL PCR tube (Corning Life Sciences, catalog number: PCR-0208-C)
4. 1.5 mL microtube (Axygen, catalog number: MCT-150-C)
5. Electroporation cuvette (Bio Rad, catalog number:1652082)
6. Polypropylene round-bottom tube (Falcon, catalog number: 352059)
7. Disposable cell spreader (BKMAMLAB, catalog number: 110305003)
8. Erlenmeyer flask (HEQI GLASS, catalog number: B-000204)
9. High-temperature resistant tissue sealing film (BKMAMLAB, catalog number: 110104005)
10. Pipette tips (KIRGEN, catalog number: KG1333)
11. Cuvettes (Loikaw, catalog number: B-002016)
12. Spectrophotometer (SHST, Model: SH-NanoOne)
Equipment
1. Pipettes (any source)
2. PCR thermocycler (Thermo Fisher Scientific, model: ProFlexTM Base)
3. DNA electrophoresis apparatus (Guangzhou Dao Yi Science and Technology Co., Ltd, model: EPHC 400)
4. Gel image analysis system (Tanon, model: MINI SPACE 960)
5. Nanodrop spectrophotometer (Shenhua Science Technology, model: SH-NnoOne)
6. Centrifuge (Eppendorf, model: 5425)
7. Bacterial incubator (Shanghai Bluepard Instruments)
8. Shaker (Shanghai Chuzhi Biotechnology, model: ZQLY-180ES)
9. Water bath (Shanghai Bluepard Instruments, model: DK-8D)
10. Electroporator (Bio-Rad, model: MicroPulser)
11. Blue-light lamp (MIULAB, model: DUT-48)
Software and datasets
1. Snapgene®4.3.6 (GSL Biotech)
2. Adobe Illustrator, 2021
3. GraphPad Prism 8
4. CHOPCHOP (https://chopchop.cbu.uib.no/)
5. TargetFinder algorithm (https://github.com/ ECBCgit/targetFinder)
Procedure
文章信息
稿件历史记录
提交日期: Aug 9, 2025
接收日期: Sep 27, 2025
在线发布日期: Oct 14, 2025
出版日期: Nov 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Yuan, X., Yu, X., Zhao, W. and Sun, J. (2025). A Practical CRISPR-Based Method for Rapid Genome Editing in Caulobacter crescentus. Bio-protocol 15(21): e5490. DOI: 10.21769/BioProtoc.5490.
分类
微生物学 > 微生物遗传学 > 基因组编辑
微生物学 > 微生物遗传学 > CRISPR-Cas9
分子生物学 > DNA > 基因表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link