- EN - English
- CN - 中文
Amplification-Free Detection of Highly Structured RNA Molecules Using SCas12aV2
发布: 2026年05月05日第16卷第9期 DOI: 10.21769/BioProtoc.5462 浏览次数: 949
评审: Marion HoggAnonymous reviewer(s)

相关实验方案

新鲜冷冻骨骼肌切片上小鼠肌肉干细胞的RNA荧光原位杂交与免疫荧光染色同步检测
Vedant R. Lakkundi [...] Albert E. Almada
2025年09月05日 1913 阅读
Abstract
The CRISPR/Cas12a system has revolutionized molecular diagnostics; however, conventional Cas12a-based methods for RNA detection typically require transcription and pre-amplification steps. Our group has recently developed a diagnostic technique known as the SCas12a assay, which combines Cas12a with a split crRNA, achieving amplification-free detection of miRNA. However, this method still encounters challenges in accurately quantifying long RNA molecules with complex secondary structures. Here, we report an enhanced version termed SCas12aV2 (split-crRNA Cas12a version 2 system), which enables direct detection of RNA molecules without sequence limitation while demonstrating high specificity in single-nucleotide polymorphism (SNP) applications. We describe the general procedure for preparing the SCas12a system and its application in detecting RNA targets from clinical samples.
Key features
• The SCas12aV2 assay enables efficient detection of long-chain RNA molecules with complex secondary structures.
• PAM-distal sites can be effectively distinguished at the SNP detection level.
• The entire experimental procedure can be completed in less than one hour.
Keywords: CRISPR/Cas12aBackground
In recent years, novel nucleic acid detection technologies, such as CRISPR/Cas12a, have been successfully applied in various fields, including viral identification, disease diagnosis, and prognostic assessment. Cas12a has two cleavage activities upon target recognition: (i) cis-cleavage, which cuts target double-stranded DNA (dsDNA) when a matching sequence is present in the crRNA spacer, and (ii) trans-cleavage, which degrades nearby single-stranded DNA (ssDNA) nonspecifically after activation. This trans-cleavage activity enables highly sensitive nucleic acid detection methods like DETECTR [1] and HOLMES [2], demonstrating their value in point-of-care diagnostics.
To date, most Cas12a-based methods [1,2] still require integration with nucleic acid amplification techniques to achieve detection efficiencies comparable to those of qPCR. Moreover, these methods are incapable of directly quantifying RNA targets. Recently, we discovered that crRNA can be divided into two fragments: scaffold RNA and spacer RNA. These fragments subsequently reassemble with the Cas12a protein to form a ribonucleoprotein (RNP) with trans-cleavage activity. Based on this discovery, we developed a split-crRNA Cas12a (SCas12a) system [3] for point-of-care testing (POCT) RNA detection technologies (Figure 1), thereby eliminating the need for reverse transcription and nucleic acid amplification. This approach achieves an average limit of detection (LoD) value of 100 fM for miRNA by fluorescence assay and of 10 fM for miRNA by lateral flow assay (LFA). Furthermore, we developed an enhanced version of the method, termed SCas12aV2 [4], which enables efficient detection of RNA substrates with long sequences and complex structures. The assay is highly versatile and can be readily adapted to various Cas12a orthologs, thus advancing molecular diagnostics by improving the accuracy and efficiency of RNA detection. Collectively, this method can be applied in various settings, such as the detection of RNA from epidemic pathogens and the identification of RNA-based cancer markers from clinical samples, facilitating rapid mutant screening for breeding purposes.
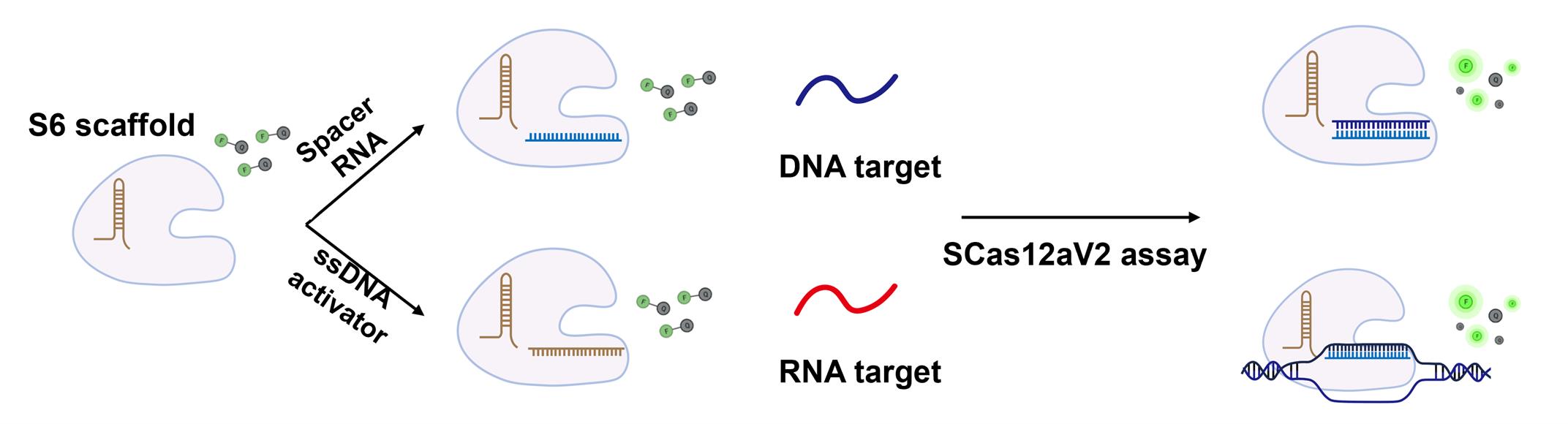
Figure 1. Schematic representation of the SCas12a assay for nucleic acid detection
Materials and reagents
Reagents
1. 10× PCR buffer (with Mg2+) (Beyotime, catalog number: D7223-1)
2. 10× NEBuffer r2.1 (NEB, catalog number: B6002S)
3. 1 M Tris pH 8.0, sterile (Sangon Biotech, catalog number: B548127-0500)
4. Sodium chloride (NaCl) (Sangon Biotech, catalog number: A610476-0001)
5. DTT (Sangon Biotech, catalog number: A620058-0100)
6. HOLMES-Fluo ssDNA reporter 1 (FAM) (TOLOBIO, catalog number: 31101), 5’FAM-TTATTATT-3’BHQ1
7. AsCas12a protein: Synthetic genes encoding the CRISPR-associated protein AsCas12a were cloned into the pET-28a(+) expression vector to construct plasmids for recombinant protein production. The sequence accuracy of these plasmids was verified prior to transformation into Escherichia coli BL21 (DE3) competent cells. For protein expression, a single colony was inoculated into LB broth supplemented with ampicillin (100 μg/mL) and incubated overnight. The following day, these cultures were diluted into 1 L of Terrific Broth (TB) medium to an optical density at 600 nm (OD600) of 0.8, cooled on ice for 10 min, induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and further incubated at 18 °C for 16 h. The bacterial cells were then pelleted by centrifugation and lysed in buffer A [20 mM Tris-HCl, pH 7.5, 300 mM NaCl, 1 mM phenylmethanesulfonyl fluoride (PMSF), 5 mM β-mercaptoethanol] containing a protease inhibitor cocktail. The recombinant Cas proteins were purified using immobilized metal affinity chromatography (IMAC) with Ni-NTA resin, followed by size-exclusion chromatography on a HiLoad® 16/600 Superdex® column according to a previously published protocol [5]. The purified proteins were concentrated using a 100 kDa molecular weight cutoff (MWCO) centrifugal filter, and their concentrations were determined by the Bradford assay. Subsequently, the concentrated proteins were flash-frozen in liquid nitrogen and stored at -80 °C for future use.
Solutions
1. 10× PCR buffer (with Mg2+) (see Recipes)
2. 10× NEBuffer r2.1 (see Recipes)
3. Storage buffer (see Recipes)
Recipes
1. 10× PCR buffer (with Mg2+)
100 mM Tris-HCl (pH 8.8 at 25 °C)
500 mM KCl
15 mM MgCl2
0.8% (v/v) Nonidet P40
2. 10× NEBuffer r2.1
500 mM NaCl
100 mM Tris-HCl
100 mM MgCl2
100 μg/mL recombinant albumin
pH 7.9 at 25 °C
3. Storage buffer
20 mM Tris-HCl
250 mM NaCl
1 mM DTT
pH 7.9 at 25 °C
Laboratory supplies
1. Pipette tips (filtered) (Thermo, catalog numbers: TF112-1000-Q, T104RS-Q, TF140-200-Q)
2. 0.2 mL PCR tubes (Thermo, catalog number: 431-MIXED-Q)
3. 1.5 mL microtubes (AXYGEN, catalog number: MCT-150-L-C)
Equipment
1. Real-time PCR system (Bio-Rad, model: CFX96 touch, or StratageneTM Mx3005P, Thermo, model: PF1457N)
2. Microcentrifuge (mySPINTM 6 Mini, Thermo, model: 75004061 or equivalent)
3. Thermal cycler (VeritiPro PCR, Thermo, model: A48141)
4. Vortex mixer (Eppendorf mixer, Eppendorf, model: 5382000074)
5. Variable volume pipettes (P10, P200, P1000)
6. Freezer (-80 °C) (Thermo, model: 905)
7. Standard laboratory equipment (analytical balance, pH meter, etc.)
Software and datasets
1. RNA secondary structure prediction software is listed as follows:
NUPACK: http://www.nupack.org/
RNAfold: http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi
Mfold: http://unafold.rna.albany.edu/?q=mfold
For the prediction of highly structured RNA regions:
1. Secondary structure prediction: Utilize NUPACK or RNAfold to identify regions with low free energy (Figure 2).
2. Target site selection: We used NUPACK to predict the secondary structures and free energy distribution of the target RNA. The results demonstrated that regions with higher free energy exhibit lower thermodynamic stability, rendering them more susceptible to disruption by activators and thus more suitable as target sites.
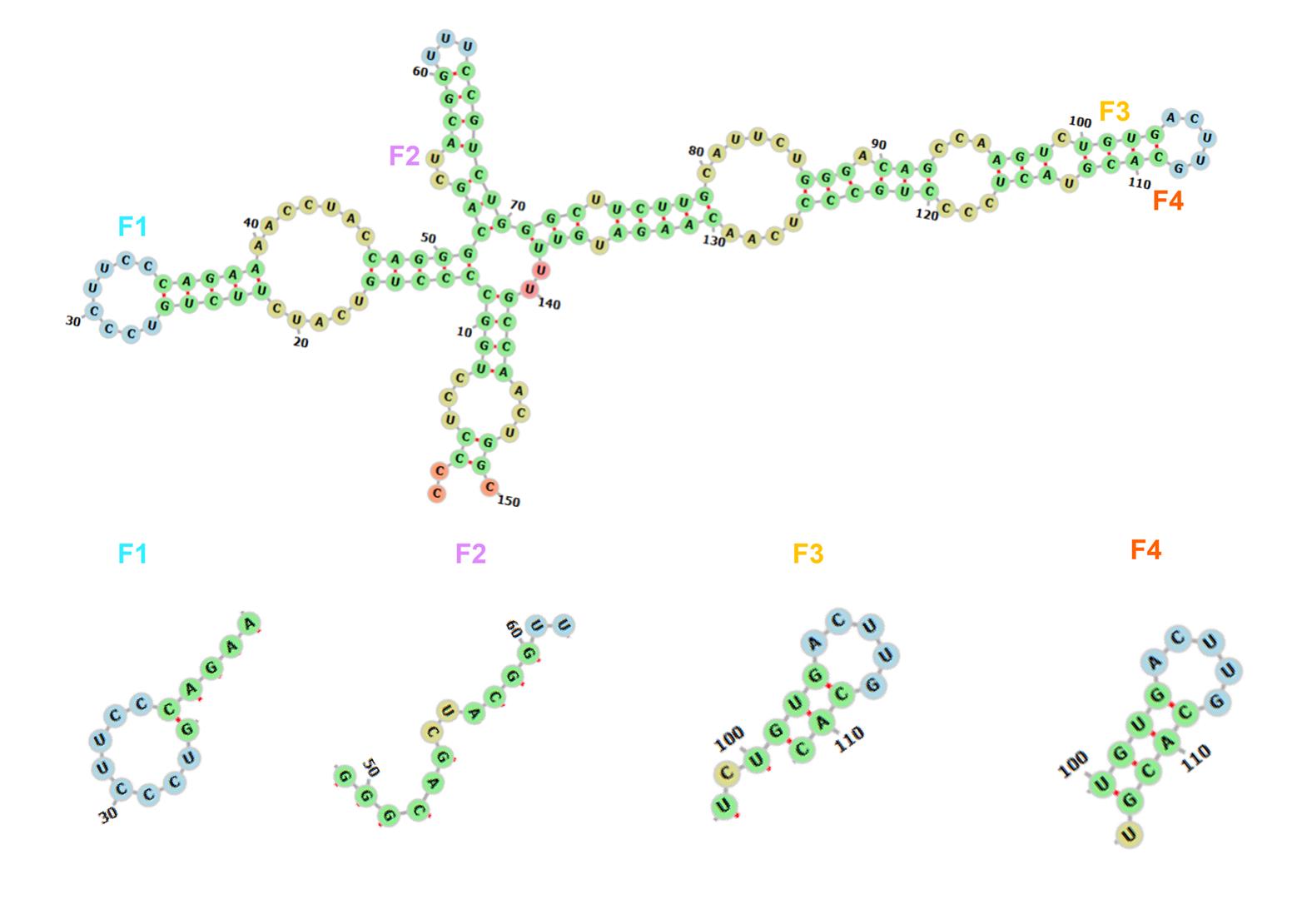
Figure 2. Schematic diagram of the structure predicted by NUPACK for the P53 RNA
For the design of the dsDNA-ssDNA hybrid activators: The hybrid activator improves detection efficiency and sensitivity compared to traditional ssDNA or dsDNA activators by enabling stronger and more stable interactions between the RNA substrate and the Cas12a active site. The design principle is outlined in the following:
1. A double-stranded DNA region: 12–16 bp for scaffold binding (Figure 3).
2. PAM sequence: TTTN (N = A, C, or G).
3. A single-stranded DNA region: 20 nt complementary to the target.
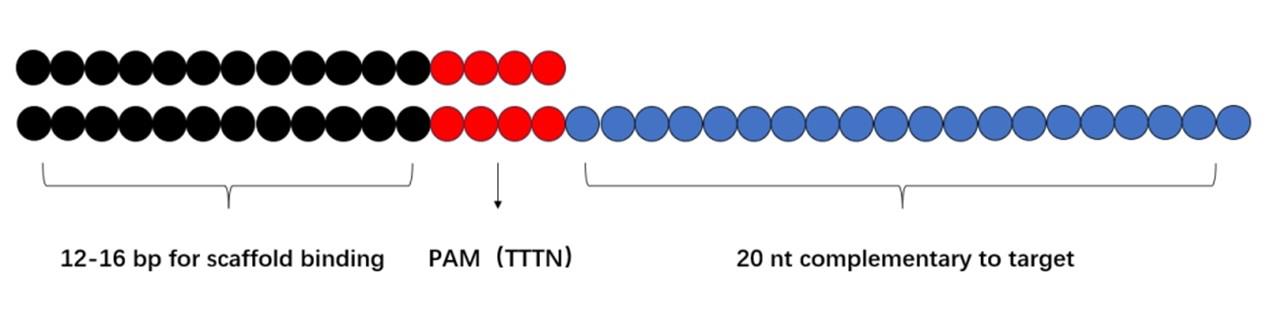
Figure 3. Schematic diagram of hybrid activator design
Procedure
文章信息
稿件历史记录
提交日期: Jun 22, 2025
接收日期: Aug 20, 2025
在线发布日期: Sep 9, 2025
出版日期: May 5, 2026
版权信息
© 2026 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Hu, T., Pei, Y., Hu, Z., Feng, J., Jiang, Q., Hu, L. and Liu, Y. (2026). Amplification-Free Detection of Highly Structured RNA Molecules Using SCas12aV2. Bio-protocol 16(9): e5462. DOI: 10.21769/BioProtoc.5462.
分类
分子生物学 > RNA > RNA 检测
微生物学 > 病原体检测 > 生物传感器
生物科学 > 生物技术 > CRISPR/Cas9
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










