- EN - English
- CN - 中文
Ultrafast Isolation of Synaptic Terminals From Rat Brain for Cryo-Electron Tomography Analysis
用于冷冻电子断层成像分析的大鼠脑突触末端超快速分离
发布: 2025年09月05日第15卷第17期 DOI: 10.21769/BioProtoc.5429 浏览次数: 3550
评审: Munenori IshibashiMario ValentinoAnonymous reviewer(s)

相关实验方案
基于微型计算机断层扫描的简化方法测量小鼠体成分、骨质疏松及肺纤维化
Madeleine B. Landau [...] Muralidharan Anbalagan
2025年02月20日 2255 阅读
Abstract
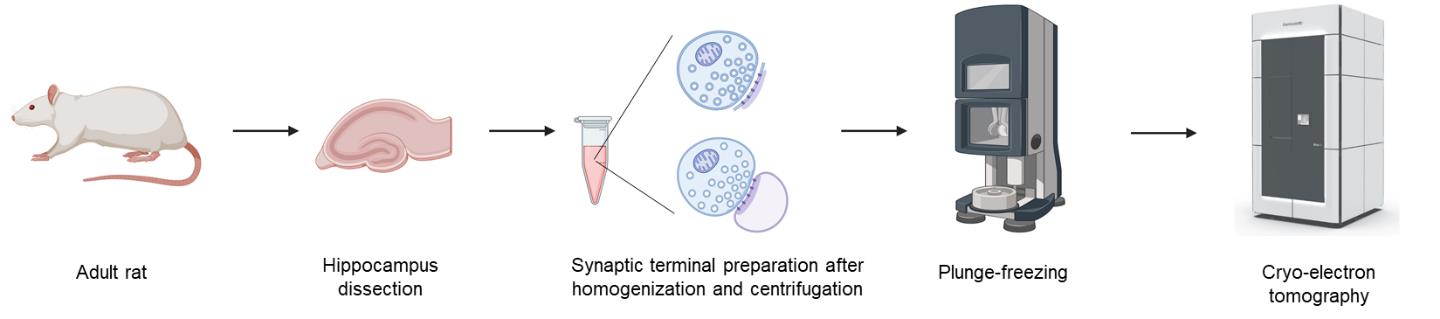
Understanding the nanoscale organization and molecular rearrangement of synaptic components is critical for elucidating the mechanisms of synaptic transmission and plasticity. Traditional synaptosome isolation protocols involve multiple centrifugation and resuspension steps, which may cause structural damage or alter the synaptosomal fraction, compromising their suitability for cryo-electron tomography (cryo-ET). Here, we present an ultrafast isolation method optimized for cryo-ET that yields two types of synaptosomal fractions: synaptosomes and synaptoneurosomes. This streamlined protocol preserves intact postsynaptic membranes apposed to presynaptic active zones and produces thin, high-quality samples suitable for in situ structural studies. The entire procedure, from tissue homogenization to vitrification, takes less than 15 min, offering a significant advantage for high-resolution cryo-ET analysis of synaptic architecture.
Key features
• Ultrafast synaptic terminal isolation from tissue homogenization to vitrification completed within 15 min.
• Retention of postsynaptic membranes with synaptic receptors and postsynaptic density (PSD) proteins.
• The thickness of the samples is suitable for in situ cryo-ET analysis.
• Enables cryo-ET studies of synaptic structures and postsynaptic membrane proteins such as AMPA receptors.
Keywords: Synaptosome (突触体)Graphical overview
Background
Cryo-electron tomography (cryo-ET) allows visualization of 3D cellular structures in their native states at nanometer resolution [1]. Synaptic elements, which are critical for neurotransmission, are particularly challenging to image with cryo-ET due to their sample thickness and molecular complexity. Isolated synaptic terminals, including synaptosomes and synaptoneurosomes, are used as a model to study synaptic structure and physiology [2,3]. Traditional methods for synaptosome or synaptoneurosome preparation often involve multiple centrifugation and resuspension steps, which might damage delicate structures and increase preparation time [4]. By streamlining the process to dissection, homogenization, and a single-step centrifugation, this protocol significantly reduces sample processing time and preserves ultrastructure, enabling direct cryo-ET studies of synaptic organelles and proteins, including synaptic vesicles, adhesion molecules, postsynaptic receptors, and synaptic nanoblocks in the postsynaptic density (PSD). Specifically, we quantitatively analyzed the distribution of tethered, docked, and partially fused synaptic vesicles, which indicates the presynaptic potential release sites, as well as the distribution of postsynaptic receptors and synaptic nanoblocks [5]. These results demonstrate that synaptic terminals isolated using our protocol are suitable for cryo-ET and subsequent quantitative analyses of synaptic components.
Materials and reagents
Biological materials
1. 10-week-old Sprague-Dawley rats of either sex (Charles River Laboratories)
Reagents
1. Isoflurane (Piramal, catalog number: NDC 66794-017-25)
2. Sucrose (Sigma-Aldrich, catalog number: S9378)
3. HEPES (Fisher BioReagent, catalog number: BP310-1)
4. EDTA (Millipore, catalog number: 324503)
5. 10-nm BSA-gold beads (Aurion, catalog number: 25486)
6. NaOH (Fisher, catalog number: AC259860010)
7. Ethane gas (A-L Compressed Gases, catalog number: AGETHANECP-7)
8. Liquid nitrogen (A-L Compressed Gases, catalog number: GSINL240)
Solutions
1. 0.5 M EDTA stock solution (see Recipes)
2. 10-nm gold beads solution (see Recipes)
3. Iso-osmotic homogenization buffer (see Recipes)
Recipes
1. 0.5 M EDTA stock solution
To prepare 0.5 M EDTA (pH 8.0), dissolve 93 g of EDTA disodium salt in ~400 mL of deionized water in a 500 mL autoclavable screw-cap bottle with a magnetic stir bar. The EDTA will not dissolve until the pH is adjusted. Slowly add ~10 g of NaOH pellets while stirring until the pH reaches 8.0. Once fully dissolved, transfer the solution to a 500 mL graduated cylinder, adjust to the final volume with deionized water, remove the stir bar, and return the solution to the bottle. Autoclave and store at room temperature.
2. 10-nm gold beads solution
To prepare a 10 nm BSA-gold solution, centrifuge 300 μL of BSA-conjugated gold nanoparticles at 16,100× g for 30 min at 4 °C. Carefully discard the supernatant without disturbing the pellet. Resuspend the pellet gently in 150 μL of iso-osmotic homogenization buffer (Recipe 3) by pipetting or brief vortexing. Store the solution at 4 °C and use within a week. 10-nm gold beads are added to the sample when plunge freezing. These gold beads will be used as fiducial markers to align the tilt series during reconstruction.
3. Iso-osmotic homogenization buffer
Prepare 500 mL of homogenization buffer as a 0.32 M sucrose solution by adding 54.77 g of sucrose in ~400 mL of deionized water and stirring in a 500 mL autoclavable screw-cap bottle with a magnetic stir bar. This is then supplemented with 0.477 g of HEPES and 1 mL of 0.5 M EDTA stock (Recipe 1) while stirring until dissolved, and finally adjusted to pH 7.4.
Laboratory supplies
1. Quantifoil R2/2 Cu 200 mesh EM grids (Quantifoil)
2. Cryo grid box (SubAngstrom, catalog number: SBV01)
Equipment
1. Small animal decapitator (Fisher Scientific, catalog number: 10-000-124)
2. Overhead stirrer (Electron Microscopy Sciences, catalog number: 6480610)
3. 10-mL Teflon head tissue grinder (Electron Microscopy Sciences, catalog number: 6479310)
4. Centrifuge (Eppendorf, model: 5415D)
5. FEI Vitrobot Mark III (FEI, now Thermo Fisher Scientific)
6. Titan Krios G4 Cryo-EM (Thermo Fisher Scientific)
7. K3 camera (Gatan)
Software and datasets
1. Thermo Fisher Tomography 5 https://www.thermofisher.com/us/en/home/electron-microscopy/products/software-em-3d-vis/tomography-software.html
2. IMOD https://bio3d.colorado.edu/imod/ [6]
3. MotionCorr2 https://emcore.ucsf.edu/ucsf-software [7]
4. Gctf https://github.com/JackZhang-Lab/GCTF [8]
Procedure
文章信息
稿件历史记录
提交日期: Jun 12, 2025
接收日期: Jul 24, 2025
在线发布日期: Aug 8, 2025
出版日期: Sep 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Sun, R. and Zhou, Q. (2025). Ultrafast Isolation of Synaptic Terminals From Rat Brain for Cryo-Electron Tomography Analysis. Bio-protocol 15(17): e5429. DOI: 10.21769/BioProtoc.5429.
- Sun, R., Allen, J. P., Mao, Z., Wilson, L., Haider, M., Alten, B., Zhou, Z., Wang, X. and Zhou, Q. (2025). The postsynaptic density in excitatory synapses is composed of clustered, heterogeneous nanoblocks. J Cell Biol. 224(6): e202406133. https://doi.org/10.1083/jcb.202406133
分类
神经科学 > 基础技术 > 快速制片
生物物理学 > 电子冷冻断层扫描 > 3D图像重建
神经科学 > 细胞机理 > 突触生理学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link