- EN - English
- CN - 中文
ClearDepth Method for Evaluations of Root Depth in Soil-Filled Pots
ClearDepth法评估土壤盆栽中根系深度
发布: 2025年08月20日第15卷第16期 DOI: 10.21769/BioProtoc.5425 浏览次数: 2116
评审: Anonymous reviewer(s)
Abstract
Despite its significant relevance to drought adaptation, optimization of nutrient acquisition, and carbon sequestration in soil, genetic factors determining root depth remain poorly explored, mostly due to the limitations of the methods currently available to estimate it. Although several such methods have been developed for crops, their applicability to large-scale studies and those involving smaller, more fragile root systems is severely limited. To address this, we have developed ClearDepth, a simple, non-destructive, low-cost method. In ClearDepth, the root system develops naturally inside the soil in clear pots. As it expands, secondary roots reach the transparent walls of the pot ("wall roots"), becoming visible. The shallowness of each wall root is then measured (wall root shallowness, WRS), and the depth of the root system is expressed as the average of all single WRS measurements. We demonstrated the suitability of ClearDepth for root depth studies using Arabidopsis thaliana and Oryza sativa (rice), two species with contrasting root system architecture (RSA) and root size. The robustness and sensitivity of the WRS trait allow us not only to reproducibly discriminate between shallow and deep root systems but also to detect smaller yet significant differences in depth determined by the influence of environmental factors, such as light. Here, we present a comprehensive protocol for utilizing this method.
Key features
• ClearDepth measures the depth of a minimum number of secondary roots, set by the user, to estimate the depth of the root system.
• The method captures differences of root depth at a spatio-developmental stage rather than at one specific time point after planting.
• ClearDepth captures differences in root depth independently of differences in total root biomass.
Keywords: Arabidopsis (拟南芥)Graphical overview
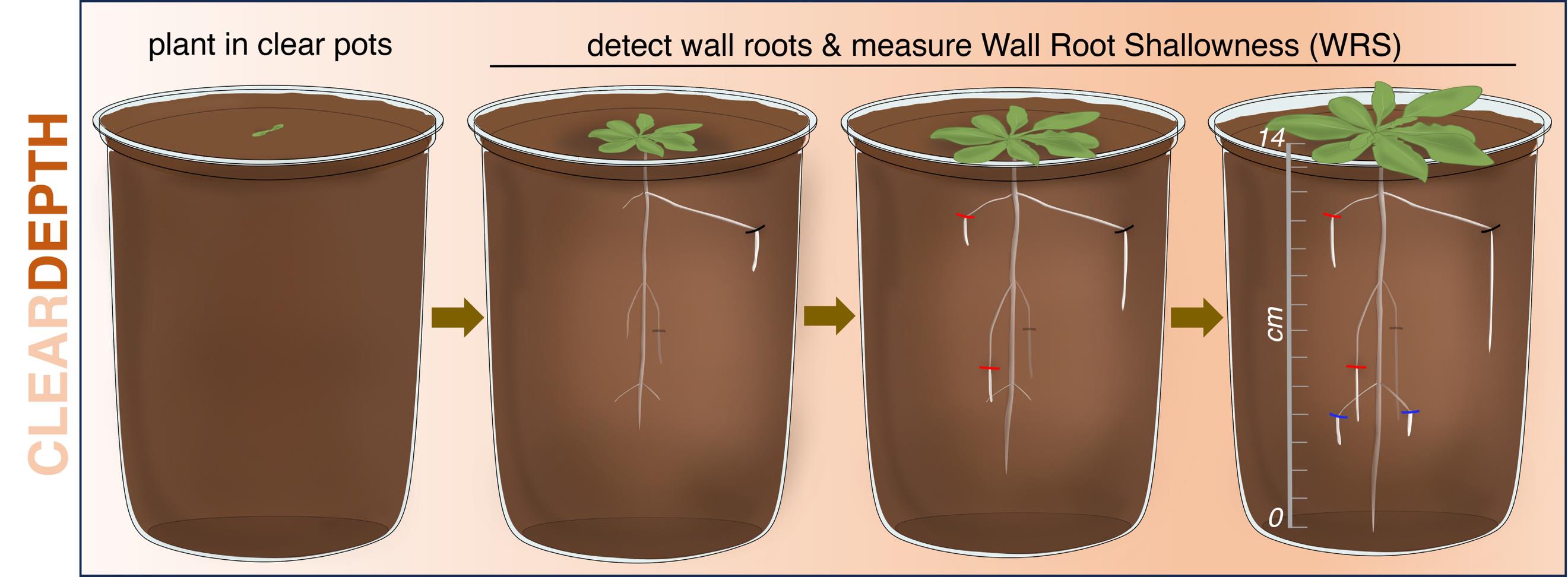
Overview of the ClearDepth method for estimations of root depth in soil. In ClearDepth, seedlings are planted in transparent pots, which allows the detection of roots growing at different depths as they touch the walls of the pot, thus becoming visible. The shallowness of individual wall roots is then measured and averaged to estimate the depth of the root system.
Background
Root depth is a trait of great relevance in the adaptation of plants to changes in the content and distribution of nutrients in the soil. Shallow roots are pivotal for the uptake of nutrients more abundantly found in topsoil layers, like phosphorus [1–3], whereas deep roots are involved in the uptake of nitrates, which move with water [4,5]. Deep roots have also been shown to improve plant resilience to drought [6–8] and are a desirable trait for strategies that aim to fight climate change by increasing carbon storage in the soil [9–11]. Despite its enormous agricultural relevance, the genetic determinants of root depth are poorly understood. This is largely due to the current lack of methods suitable for mid- and large-scale studies of root depth in soil. While excavation methods (shovelomics) and imaging techniques like X-ray have proved useful to estimate the distribution of root biomass across soil depths in crop species, the amount of labor, the complexity of image analysis, the destructiveness of the methods, and their cost, among other factors, severely limit their use to rather small, validation experiments [12–17]. Such limitations are even more severe when the study involves species characterized by small, fragile roots like the model organism Arabidopsis. In recent years, at least two methods for the study of the Arabidopsis root system architecture (RSA) and root depth in soil have been published. The GLO-Roots method relies on the use of plants transformed to express the luciferase marker, which allows the imaging of roots growing in thin, soil-filled, transparent pots [18]. The GLO-RIA software is then used to analyze the images and extract traits such as depth [18]. In Ogura's method, whole pots containing the root system are sectioned in two halves at the center of the plant, and images of the exposed roots are obtained and processed for measurements of "shallowness" (standard deviation of all root pixels in x-axis direction in a picture divided by that in y-axis direction), a proxy for root depth [19]. Although both provide valuable insights into the genetic control of RSA traits and root depth in Arabidopsis, their applicability is restricted by the need to use transformants, in one case, and the considerable amount of labor, in the other.
We have recently developed ClearDepth, a conceptually simple, low-cost, and robust method to evaluate root depth in soil [20]. The method relies on the use of clear pots, allowing the visualization of roots that reach the transparent walls of the pot ("wall roots") after having grown buried in the soil. Importantly, in our method, the root system develops entirely inside the soil, in a three-dimensional, natural manner. This contrasts with other methods that rely on the use of rhizotrons, where the roots grow in a restricted, almost 2D soil environment that severely limits their radial expansion. The shallowness of each wall root (wall root shallowness, WRS) is measured, and the depth of the root system is expressed as the average of all single WRS measurements (mean WRS). Deep-rooted and shallow-rooted systems are characterized by relatively low or high mean WRS values, respectively.
The mean WRS parameter captures differences of root depth at a developmental stage defined by a minimum number of wall roots or WRS measurements set by the user (in our case, five; Arabidopsis and rice), and not at a specific time after planting. Importantly, estimates of root depth by the mean WRS parameter are independent of total root biomass, and the method does not measure root length distribution by depth, typical of, for example, shovelomics-based approaches. ClearDepth's second trait, resolution time (RT), considers the temporal aspect of root system expansion in the soil. It is defined as the time needed for a root system to display the minimum number of wall roots established by the user. Although less sensitive than the mean WRS parameter to differentiate between shallow and deep root systems, RT can be especially useful to compare root systems that do not differ significantly in mean WRS: the faster the wall roots appear in deep soil layers, the deeper the root system.
The value of ClearDepth is not limited to estimations of Arabidopsis' root depth; the method can also be applied to species characterized by larger, fibrous root systems, like the monocot rice. In addition, the method not only distinguishes between shallow-rooted and deep-rooted systems but is sensitive enough to detect small differences in root depth determined by the influence of, for example, an environmental factor like light. ClearDepth’s main limitation is the relatively long times (30–45 days) that some deep-rooted systems require to display wall roots. This and other aspects of the method, such as the automation of WRS measurements and the extension of its use to species other than Arabidopsis and rice, are current areas of improvement and exploration. In conclusion, ClearDepth offers a simple, low-cost, and easily scalable method for robust and sensitive measurements of root depth in soil-filled pots in controlled environment settings. Here, we provide a detailed protocol for its use.
Materials and reagents
Biological materials
1. Seeds of the accessions, varieties, mutants or genotypes to evaluate
Reagents
1. Murashige and Skoog (MS) basal salts (Caisson, catalog number: MSP01-1KG)
2. Sucrose (Fisher Bioreagents, catalog number: BP220-10)
3. 1 M KOH (Sigma-Aldrich, catalog number: 244720)
3. Phytagel (plantMedia, catalog number: 40000035-3)
4. Germicidal ultra bleach (Waxie, catalog number: 170018)
5. 1 N HCl (Fisher Scientific, catalog number: SA48-1)
6. Sterile (autoclaved) ddH2O
Solutions
1. 1/4 strength MS phytagel medium (see Recipes)
Recipes
1. 1/4 strength MS phytagel medium
| Reagent | Final concentration | Quantity or Volume |
| MS basal salts | 1/4 strength | 1.1 g |
| Sucrose | 1% | 10 g |
| Phytagel | 1% | 10 g |
| ddH2O | n/a | up to 1 L |
Laboratory supplies
1. 14 × 9–10.7 cm (depth × diameter) pots (Webrestaurant Store) (https://www.webstaurantstore.com/choice-32-oz- microwavable-contact-clear-round-deli-container-pack/999RD32BULK.html)
2. 12 × 12 cm plates (Greiner Bio-One, catalog number: 688161)
3. 10 × 15 inch seed germination paper (Nasco Education, catalog number: SB39211)
4. Sunshine Mix 4 soil (Sungro, https://www.sungro.com/retail-product/sunshine-mix-4-aggregate-plus-with-mycorrhizae) containing a mix of Canadian sphagnum peat moss, perlite, dolomite lime, a proprietary blend of endomycorrhiza, RESiLIENCE® for improved resistance to wilting, a wetting agent, and a low phosphorus fertilizer
5. Sterile 100 mL serological pipettes (Corning Incorporated, catalog number: 4484)
6. Graduated beakers (Grainger, catalog number: 3VEZ3)
7. 1 L graduated Pyrex bottles (Sigma-Aldrich, catalog number: CLS13951L)
8. 3M surgical tape (Micropore, catalog number: 1530-0)
9. FisherbrandTM fine precision medium tipped tweezers/forceps (Fisher Scientific, catalog number: 12-000-157)
10. 250 mL graduated Pyrex beakers (Sigma-Aldrich, catalog number: CLS1000250)
11. 2 mL microcentrifuge tubes (SealRite, catalog number: 1620-2700)
12. 96-well microtube rack (Sigma-Aldrich, catalog number: 8780)
13. Sterile 200 μL pipette tips (USA Scientific TipOne®, catalog number: 1110-1730)
Equipment
1. Laminar flow hood (NuAire, model: Nu.425.600)
2. Analytical balance (Bonvoisin, model: HZ50002B)
3. pH meter (Fisher Scientific, model: Accumet AB150)
4. Magnetic stir plate (Fisher Scientific, model: Isotemp) and magnetic stir bars (Fisher Scientific)
5. Pipetboy (VWR, model: Powerpette Plus)
6. Autoclave (Consolidated Sterilizer Systems, CSS, model: 3AV-5)
7. Walk-in growth chamber (Conviron, model: MTPS216) or reach-in vertical growth chamber (Conviron, model: PGC20)
8. Fume Hood (Hanson)
9. NalgeneTM bio-transport carrier (Thermo Scientific, catalog number: 7135-0001)
10. Microwave Oven (Panasonic)
11. Fridge or cold room (Imperial Manufacturing)
12. Water bath (Thermo Fisher, model: Precision 50)
Procedure
文章信息
稿件历史记录
提交日期: May 12, 2025
接收日期: Jul 10, 2025
在线发布日期: Aug 5, 2025
出版日期: Aug 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Rosquete, M. R., Gonzalez, J., Gonzalez, N., Wertz, K., Patil, S. and Busch, W. (2025). ClearDepth Method for Evaluations of Root Depth in Soil-Filled Pots. Bio-protocol 15(16): e5425. DOI: 10.21769/BioProtoc.5425.
分类
植物科学 > 植物生理学 > 表型分析
植物科学 > 植物发育生物学 > 向地性
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link